Gene
KWMTBOMO13770 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011479
Annotation
PREDICTED:_cytosolic_purine_5'-nucleotidase_isoform_X2_[Plutella_xylostella]
Location in the cell
Cytoplasmic Reliability : 2.581
Sequence
CDS
ATGGATGTTTGTAAGGAAAACGAGTGCCTGACTCTTCAAGATAAAAATATTATGTCCAAGAAATACTATCGTCGTGTTGAACACAGAATTTTCGTTAACAGATCGTTGCATTTAGAAAATGTTAAATTTTACGGTTTTGATATGGACTATACATTGGCCGAATACAAATCGCCACAGTACGAAACATTAGGATTCAATTTAACCAAAGAAAGATTAGTAGTCAAAGGATATCCAAGAGAGATATTAGAATTCGAATATGACCCTTCGTTTCCAGTGAGAGGTCTCTGGTTTGATACGTTATATGGAAATTTGCTCAAAGTCGATGCCTACGGGAATATTTTAGTGTGTGTACATGGATTTGAATTTTTAAAACATTCACAGGTATACGAATTATATCCGAATAAATTCTTGACTTTAGATGAATCGAGGGTTTACGTTCTGAACACTTTGTTTAACCTGCCGGAAACCTATCTGATTGCGTGCCTCATAGATTTCTTCACGAATTCACCCCAATATACGAGAGTTAAGACGGGAGTGAGATGCGGCGACCTAGCTATGTCCTTCAAATCAATCTTCCAAGATGTGCGAAATGCCGTCGACTACGTTCACATACACGGCGATCTCAAGAAGAAAACAACGGAAAACTTAGATTTGTACCTGAAGAAAGACGAAAGACTGCCAATGTTCTTGTCTAGGATAAGAGAAAGCGGAGCTAGACTGTTTATACTAACTAATAGCGATTACAATTTCACTGATAAAATCATGAATTACCTATTCGATTTCCCGCACGGTGCTAAGGTCGGTGAGCCCCACCGGAACTGGAAGACTTATTTCGACTGGATCGTAGTGGACGCTAAGAAGCCTCTGTTCTTCGGCGAGGGAACCACTCTAAGGCAGGTCGACACTCGCACCGGCGCTCTGAAGATGGGACATCACGTAGGACCACTGCACGAGGGATCCGTCTATTCAGGAGGCTCGTGTGACGTATTCACGGAGCTGATCAGCGCCAAAGGCAAAGACGTTTTGTACATCGGCGATCACATATTCGGGGACATCCTCAAATCGAAGAAAATCGGCGGCTGGCGAACCTTCCTCGTTGTGCCCGAATTGGTTCAGGAGCTCCACGTGTGGACCGACAAGTGCCAGTTGTTCGCGCAGCTGCAGACCCTCGACGTTCAGCTCGGCGATATGTACAAAGACTTGGATTCGAGTACAAAAGAGAAACCGGATATATCCAAGCTCCGCATGTCGATACGCGACGTGACGCACAAGATGGATCTGGCGTACGGCATGCTCGGCTCGCTATTCCGTTCCGGTTCGAGACAAACCTTCTTCTCATCTCAGGTGGTGAGGTACGCCGATCTGTACGCGGCTTCGTTCCTCAACTTGATGTACTATCCGTTCTGCTACATGTTCCGTGCGCCCGCCATGCTGATGCCGCACGAGTCTACGGTGGCGCACGAACAACGGTTCACCCTCGACACGCCGACCATTGACCGATCTAGACATCCAAGCGCACAACGTAGCTATGGCGTCGTCACTGTAGCGGAGGACACTTCTGAAGAAAACAGCATCATCAACGCACAAGAACCCATTGATGCCAAAGCAGCAGGATCCGTGCCCCACGTACGGCCTGAAACACCCCGACAACTCACGCACACACACGACGACGACTGTTCGGACGACGAAGACGTCACAAAGGCGGCGCAACACAAATAA
Protein
MDVCKENECLTLQDKNIMSKKYYRRVEHRIFVNRSLHLENVKFYGFDMDYTLAEYKSPQYETLGFNLTKERLVVKGYPREILEFEYDPSFPVRGLWFDTLYGNLLKVDAYGNILVCVHGFEFLKHSQVYELYPNKFLTLDESRVYVLNTLFNLPETYLIACLIDFFTNSPQYTRVKTGVRCGDLAMSFKSIFQDVRNAVDYVHIHGDLKKKTTENLDLYLKKDERLPMFLSRIRESGARLFILTNSDYNFTDKIMNYLFDFPHGAKVGEPHRNWKTYFDWIVVDAKKPLFFGEGTTLRQVDTRTGALKMGHHVGPLHEGSVYSGGSCDVFTELISAKGKDVLYIGDHIFGDILKSKKIGGWRTFLVVPELVQELHVWTDKCQLFAQLQTLDVQLGDMYKDLDSSTKEKPDISKLRMSIRDVTHKMDLAYGMLGSLFRSGSRQTFFSSQVVRYADLYAASFLNLMYYPFCYMFRAPAMLMPHESTVAHEQRFTLDTPTIDRSRHPSAQRSYGVVTVAEDTSEENSIINAQEPIDAKAAGSVPHVRPETPRQLTHTHDDDCSDDEDVTKAAQHK
Summary
Cofactor
Mg(2+)
Uniprot
H9JPM4
A0A1E1WPX9
A0A1E1WPL4
A0A3S2M6B7
A0A212FEB1
A0A2W1BIS0
+ More
A0A194RUL9 A0A2A4J758 A0A3L8DS42 A0A158NKF0 A0A151I8W5 A0A151WTL4 A0A195EJI9 A0A195BU80 A0A195F5U7 E2C3D3 A0A411G761 A0A2J7PCP8 A0A2A3EF98 A0A0L0C0R1 V9IC03 A0A1B6C1W6 A0A1I8PDF4 K7IXY4 A0A1I8PDF9 A0A1B6DUW2 A0A232EM26 U5EKI4 A0A310SF18 A0A1I8PDF0 A0A2J7PCR4 A0A067RTV1 A0A1Q3FV94 A0A1B6CJB5 A0A0L7RJK2 A0A0M8ZZF1 A0A1B6I6W2 A0A1L8DXT2 A0A1L8DXP4 A0A1B6FES8 A0A1B6EKK5 A0A1I8MZL2 A0A1B6HUF9 A0A088ADD3 A0A0V0G5E5 F5HJK7 A0A1I8MZL8 A0A1I8MZL6 A0A1Q3FUW5 V9I9R1 A0A1Q3FV02 A0A1Q3FV16 A0A1Q3FUY5 A0A224XEG9 A0A1Q3FVE7 A0A1I8PDF5 A0A1Q3FUV6 A0A1I8PD53 A0A1B6DS89 A0A1I8MZL3 A0A1I8PD72 A0A182IAX6 A0A1Y9IZ76 A0A023F4N3 A0A0N7Z8J6 A0A161TF96 A0A1I8MZL0 A0A1S4G8S1 A0A2M4BI06 A0A146LNW5 A0A2M3ZGE0 T1I697 A0A182W5Q3 A0A2M3ZGM0 T1PEI3 A0A2M4BGR9 A0A182XJZ6 A0A1B6JI44 A0A069DV38 A0A182LWW2 A0A1I8PDC8 A0A182YCV4 A0A1B6IVB5 A0A182V577 A0A2M4A9E6 A0A182N8I3 A0A2M3Z286 Q7QFA2 A0A1I8MZL1 E0VIF5 A0A1B6C9T9 A0A084VV76 A0A1L8DXP6 A0A146LDT8 A0A2M4A8Q2 A0A182QBV1 A0A182RHN6 A0A0K8W227 W8AHP5 W5JSV7
A0A194RUL9 A0A2A4J758 A0A3L8DS42 A0A158NKF0 A0A151I8W5 A0A151WTL4 A0A195EJI9 A0A195BU80 A0A195F5U7 E2C3D3 A0A411G761 A0A2J7PCP8 A0A2A3EF98 A0A0L0C0R1 V9IC03 A0A1B6C1W6 A0A1I8PDF4 K7IXY4 A0A1I8PDF9 A0A1B6DUW2 A0A232EM26 U5EKI4 A0A310SF18 A0A1I8PDF0 A0A2J7PCR4 A0A067RTV1 A0A1Q3FV94 A0A1B6CJB5 A0A0L7RJK2 A0A0M8ZZF1 A0A1B6I6W2 A0A1L8DXT2 A0A1L8DXP4 A0A1B6FES8 A0A1B6EKK5 A0A1I8MZL2 A0A1B6HUF9 A0A088ADD3 A0A0V0G5E5 F5HJK7 A0A1I8MZL8 A0A1I8MZL6 A0A1Q3FUW5 V9I9R1 A0A1Q3FV02 A0A1Q3FV16 A0A1Q3FUY5 A0A224XEG9 A0A1Q3FVE7 A0A1I8PDF5 A0A1Q3FUV6 A0A1I8PD53 A0A1B6DS89 A0A1I8MZL3 A0A1I8PD72 A0A182IAX6 A0A1Y9IZ76 A0A023F4N3 A0A0N7Z8J6 A0A161TF96 A0A1I8MZL0 A0A1S4G8S1 A0A2M4BI06 A0A146LNW5 A0A2M3ZGE0 T1I697 A0A182W5Q3 A0A2M3ZGM0 T1PEI3 A0A2M4BGR9 A0A182XJZ6 A0A1B6JI44 A0A069DV38 A0A182LWW2 A0A1I8PDC8 A0A182YCV4 A0A1B6IVB5 A0A182V577 A0A2M4A9E6 A0A182N8I3 A0A2M3Z286 Q7QFA2 A0A1I8MZL1 E0VIF5 A0A1B6C9T9 A0A084VV76 A0A1L8DXP6 A0A146LDT8 A0A2M4A8Q2 A0A182QBV1 A0A182RHN6 A0A0K8W227 W8AHP5 W5JSV7
Pubmed
EMBL
BABH01015247
BABH01015248
GDQN01005664
GDQN01001985
JAT85390.1
JAT89069.1
+ More
GDQN01011734 GDQN01002278 JAT79320.1 JAT88776.1 RSAL01000022 RVE52395.1 AGBW02008970 OWR52027.1 KZ150160 PZC72726.1 KQ459833 KPJ19836.1 NWSH01002927 PCG67280.1 QOIP01000005 RLU22679.1 ADTU01002062 ADTU01002063 ADTU01002064 KQ978331 KYM95067.1 KQ982757 KYQ51107.1 KQ978801 KYN28301.1 KQ976403 KYM92132.1 KQ981805 KYN35552.1 GL452320 EFN77471.1 MH365904 QBB01713.1 NEVH01027057 PNF14118.1 KZ288271 PBC29826.1 JRES01001065 KNC25875.1 JR037574 AEY57854.1 GEDC01029944 JAS07354.1 GEDC01007835 JAS29463.1 NNAY01003442 OXU19410.1 GANO01001802 JAB58069.1 KQ760549 OAD59881.1 PNF14120.1 KK852468 KDR23274.1 GFDL01003649 JAV31396.1 GEDC01023761 JAS13537.1 KQ414581 KOC71014.1 KQ435812 KOX72743.1 GECU01025047 JAS82659.1 GFDF01002865 JAV11219.1 GFDF01002866 JAV11218.1 GECZ01021072 JAS48697.1 GECZ01031305 JAS38464.1 GECU01029406 JAS78300.1 GECL01002862 JAP03262.1 AAAB01008846 EGK96468.1 GFDL01003646 JAV31399.1 JR037572 JR037573 AEY57852.1 AEY57853.1 GFDL01003648 JAV31397.1 GFDL01003650 JAV31395.1 GFDL01003651 JAV31394.1 GFTR01007032 JAW09394.1 GFDL01003652 JAV31393.1 GFDL01003656 JAV31389.1 GEDC01008752 JAS28546.1 APCN01001357 APCN01001358 GBBI01002365 JAC16347.1 GDKW01003163 JAI53432.1 GEMB01001025 JAS02122.1 GGFJ01003482 MBW52623.1 GDHC01010173 JAQ08456.1 GGFM01006886 MBW27637.1 ACPB03001800 GGFM01006890 MBW27641.1 KA647197 AFP61826.1 GGFJ01003010 MBW52151.1 GECU01008848 JAS98858.1 GBGD01001099 JAC87790.1 AXCM01001073 GECU01016825 JAS90881.1 GGFK01004105 MBW37426.1 GGFM01001879 MBW22630.1 EAA06405.5 DS235199 EEB13161.1 GEDC01027055 JAS10243.1 ATLV01017157 KE525157 KFB41870.1 GFDF01002861 JAV11223.1 GDHC01012830 JAQ05799.1 GGFK01003863 MBW37184.1 AXCN02002231 GDHF01029740 GDHF01014685 GDHF01007183 JAI22574.1 JAI37629.1 JAI45131.1 GAMC01021368 JAB85187.1 ADMH02000256 ETN67226.1
GDQN01011734 GDQN01002278 JAT79320.1 JAT88776.1 RSAL01000022 RVE52395.1 AGBW02008970 OWR52027.1 KZ150160 PZC72726.1 KQ459833 KPJ19836.1 NWSH01002927 PCG67280.1 QOIP01000005 RLU22679.1 ADTU01002062 ADTU01002063 ADTU01002064 KQ978331 KYM95067.1 KQ982757 KYQ51107.1 KQ978801 KYN28301.1 KQ976403 KYM92132.1 KQ981805 KYN35552.1 GL452320 EFN77471.1 MH365904 QBB01713.1 NEVH01027057 PNF14118.1 KZ288271 PBC29826.1 JRES01001065 KNC25875.1 JR037574 AEY57854.1 GEDC01029944 JAS07354.1 GEDC01007835 JAS29463.1 NNAY01003442 OXU19410.1 GANO01001802 JAB58069.1 KQ760549 OAD59881.1 PNF14120.1 KK852468 KDR23274.1 GFDL01003649 JAV31396.1 GEDC01023761 JAS13537.1 KQ414581 KOC71014.1 KQ435812 KOX72743.1 GECU01025047 JAS82659.1 GFDF01002865 JAV11219.1 GFDF01002866 JAV11218.1 GECZ01021072 JAS48697.1 GECZ01031305 JAS38464.1 GECU01029406 JAS78300.1 GECL01002862 JAP03262.1 AAAB01008846 EGK96468.1 GFDL01003646 JAV31399.1 JR037572 JR037573 AEY57852.1 AEY57853.1 GFDL01003648 JAV31397.1 GFDL01003650 JAV31395.1 GFDL01003651 JAV31394.1 GFTR01007032 JAW09394.1 GFDL01003652 JAV31393.1 GFDL01003656 JAV31389.1 GEDC01008752 JAS28546.1 APCN01001357 APCN01001358 GBBI01002365 JAC16347.1 GDKW01003163 JAI53432.1 GEMB01001025 JAS02122.1 GGFJ01003482 MBW52623.1 GDHC01010173 JAQ08456.1 GGFM01006886 MBW27637.1 ACPB03001800 GGFM01006890 MBW27641.1 KA647197 AFP61826.1 GGFJ01003010 MBW52151.1 GECU01008848 JAS98858.1 GBGD01001099 JAC87790.1 AXCM01001073 GECU01016825 JAS90881.1 GGFK01004105 MBW37426.1 GGFM01001879 MBW22630.1 EAA06405.5 DS235199 EEB13161.1 GEDC01027055 JAS10243.1 ATLV01017157 KE525157 KFB41870.1 GFDF01002861 JAV11223.1 GDHC01012830 JAQ05799.1 GGFK01003863 MBW37184.1 AXCN02002231 GDHF01029740 GDHF01014685 GDHF01007183 JAI22574.1 JAI37629.1 JAI45131.1 GAMC01021368 JAB85187.1 ADMH02000256 ETN67226.1
Proteomes
UP000005204
UP000283053
UP000007151
UP000053240
UP000218220
UP000279307
+ More
UP000005205 UP000078542 UP000075809 UP000078492 UP000078540 UP000078541 UP000008237 UP000235965 UP000242457 UP000037069 UP000095300 UP000002358 UP000215335 UP000027135 UP000053825 UP000053105 UP000095301 UP000005203 UP000007062 UP000075840 UP000076407 UP000015103 UP000075920 UP000075883 UP000076408 UP000075903 UP000075884 UP000009046 UP000030765 UP000075886 UP000075900 UP000000673
UP000005205 UP000078542 UP000075809 UP000078492 UP000078540 UP000078541 UP000008237 UP000235965 UP000242457 UP000037069 UP000095300 UP000002358 UP000215335 UP000027135 UP000053825 UP000053105 UP000095301 UP000005203 UP000007062 UP000075840 UP000076407 UP000015103 UP000075920 UP000075883 UP000076408 UP000075903 UP000075884 UP000009046 UP000030765 UP000075886 UP000075900 UP000000673
PRIDE
Pfam
PF05761 5_nucleotid
Interpro
SUPFAM
SSF56784
SSF56784
Gene 3D
ProteinModelPortal
H9JPM4
A0A1E1WPX9
A0A1E1WPL4
A0A3S2M6B7
A0A212FEB1
A0A2W1BIS0
+ More
A0A194RUL9 A0A2A4J758 A0A3L8DS42 A0A158NKF0 A0A151I8W5 A0A151WTL4 A0A195EJI9 A0A195BU80 A0A195F5U7 E2C3D3 A0A411G761 A0A2J7PCP8 A0A2A3EF98 A0A0L0C0R1 V9IC03 A0A1B6C1W6 A0A1I8PDF4 K7IXY4 A0A1I8PDF9 A0A1B6DUW2 A0A232EM26 U5EKI4 A0A310SF18 A0A1I8PDF0 A0A2J7PCR4 A0A067RTV1 A0A1Q3FV94 A0A1B6CJB5 A0A0L7RJK2 A0A0M8ZZF1 A0A1B6I6W2 A0A1L8DXT2 A0A1L8DXP4 A0A1B6FES8 A0A1B6EKK5 A0A1I8MZL2 A0A1B6HUF9 A0A088ADD3 A0A0V0G5E5 F5HJK7 A0A1I8MZL8 A0A1I8MZL6 A0A1Q3FUW5 V9I9R1 A0A1Q3FV02 A0A1Q3FV16 A0A1Q3FUY5 A0A224XEG9 A0A1Q3FVE7 A0A1I8PDF5 A0A1Q3FUV6 A0A1I8PD53 A0A1B6DS89 A0A1I8MZL3 A0A1I8PD72 A0A182IAX6 A0A1Y9IZ76 A0A023F4N3 A0A0N7Z8J6 A0A161TF96 A0A1I8MZL0 A0A1S4G8S1 A0A2M4BI06 A0A146LNW5 A0A2M3ZGE0 T1I697 A0A182W5Q3 A0A2M3ZGM0 T1PEI3 A0A2M4BGR9 A0A182XJZ6 A0A1B6JI44 A0A069DV38 A0A182LWW2 A0A1I8PDC8 A0A182YCV4 A0A1B6IVB5 A0A182V577 A0A2M4A9E6 A0A182N8I3 A0A2M3Z286 Q7QFA2 A0A1I8MZL1 E0VIF5 A0A1B6C9T9 A0A084VV76 A0A1L8DXP6 A0A146LDT8 A0A2M4A8Q2 A0A182QBV1 A0A182RHN6 A0A0K8W227 W8AHP5 W5JSV7
A0A194RUL9 A0A2A4J758 A0A3L8DS42 A0A158NKF0 A0A151I8W5 A0A151WTL4 A0A195EJI9 A0A195BU80 A0A195F5U7 E2C3D3 A0A411G761 A0A2J7PCP8 A0A2A3EF98 A0A0L0C0R1 V9IC03 A0A1B6C1W6 A0A1I8PDF4 K7IXY4 A0A1I8PDF9 A0A1B6DUW2 A0A232EM26 U5EKI4 A0A310SF18 A0A1I8PDF0 A0A2J7PCR4 A0A067RTV1 A0A1Q3FV94 A0A1B6CJB5 A0A0L7RJK2 A0A0M8ZZF1 A0A1B6I6W2 A0A1L8DXT2 A0A1L8DXP4 A0A1B6FES8 A0A1B6EKK5 A0A1I8MZL2 A0A1B6HUF9 A0A088ADD3 A0A0V0G5E5 F5HJK7 A0A1I8MZL8 A0A1I8MZL6 A0A1Q3FUW5 V9I9R1 A0A1Q3FV02 A0A1Q3FV16 A0A1Q3FUY5 A0A224XEG9 A0A1Q3FVE7 A0A1I8PDF5 A0A1Q3FUV6 A0A1I8PD53 A0A1B6DS89 A0A1I8MZL3 A0A1I8PD72 A0A182IAX6 A0A1Y9IZ76 A0A023F4N3 A0A0N7Z8J6 A0A161TF96 A0A1I8MZL0 A0A1S4G8S1 A0A2M4BI06 A0A146LNW5 A0A2M3ZGE0 T1I697 A0A182W5Q3 A0A2M3ZGM0 T1PEI3 A0A2M4BGR9 A0A182XJZ6 A0A1B6JI44 A0A069DV38 A0A182LWW2 A0A1I8PDC8 A0A182YCV4 A0A1B6IVB5 A0A182V577 A0A2M4A9E6 A0A182N8I3 A0A2M3Z286 Q7QFA2 A0A1I8MZL1 E0VIF5 A0A1B6C9T9 A0A084VV76 A0A1L8DXP6 A0A146LDT8 A0A2M4A8Q2 A0A182QBV1 A0A182RHN6 A0A0K8W227 W8AHP5 W5JSV7
PDB
6FXH
E-value=0,
Score=1754
Ontologies
PATHWAY
PANTHER
Topology
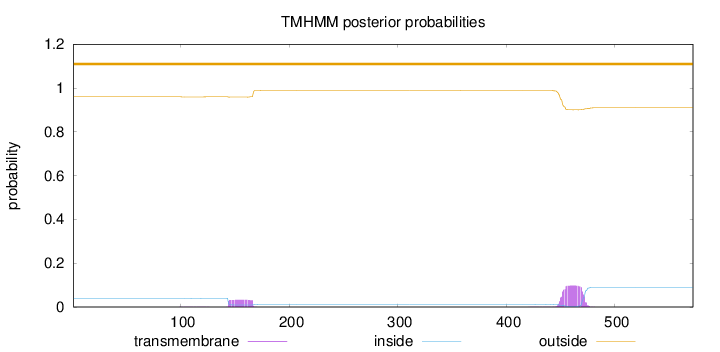
Length:
572
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
2.81153
Exp number, first 60 AAs:
0.00121
Total prob of N-in:
0.04027
outside
1 - 572
Population Genetic Test Statistics
Pi
256.078852
Theta
186.564507
Tajima's D
1.130126
CLR
0.353898
CSRT
0.691465426728664
Interpretation
Uncertain
