Pre Gene Modal
BGIBMGA011480
Annotation
PREDICTED:_ribonuclease_H2_subunit_A-like_[Plutella_xylostella]
Full name
Ribonuclease
Location in the cell
Nuclear Reliability : 1.525
Sequence
CDS
ATGAACTCACTACAAGAATTACAGGCAATTATTAAATCCAAAGATAATTTCACTAAATTTGTCATAAATTCTGAAGTACCGGCTGTCTGCAAAAATGAACCTTGCATGCTCGGAGTGGATGAAGCAGGCCGTGGTCCTGTGCTCGGACCTATGGTGTATGGAGTATCCTACTGCCCTCTAAATCGAAAGAGTATTTTGGAGGAATTGGGCTGTGCTGACTCTAAAGCTCTTACGGAAGCTAAAAGGGATGAAATATTCACAAAGATGTTGACTGAGGAAGACAGCCTCAATAATGTTGGCTGGGCTGTTGAAGTCATATCACCTAATTACATTTCCAATTCAATGTACAGACGAGCCAAATGTTCATTAAATGAAGTGTCAATGAATTCTGCAATAGATTTAATTAAGAAAGCTGCAGAAAGTGGTGTCAATATTACAGAAGTTTATGTGGACACAGTGGGGCCTCCAGAGAAATATCAAACAAAATTATCTGAAATATTTCCTTCTTACAAAATAACTGTGGCGAAAAAGGCCGACTCCATCTATCCCATAGTATCAGCAGCAAGTATTGTGGCTAAAGTAACCAGAGATCATGCACTAAAAGTATGGCATTTTAATGAGGGACTACAATTGGATCACACAGAATTTGGCAGTGGATATCCGGGAGATCCATTAACAAAGAAATTCATACGCGATCGAATCGATAACGTTTTTGGATATCCGCTTTTGGTACGGTTCAGCTGGTCGACAGCAGAACTCATGCTGCAAGAGAAAGCGGCAGCGTGCACGTTTGAAGACGTAGACGACCCTGGCAAAGGAAAAATCGCGATCGATGGCGATAACTTCCTTCGGGCGGTAGCTAAATTTACAATCGGCAAAATGTTGTTTCGGTATTGTGCTGCTTTACTGGCGCTCGCCGTTTGCGCCTCCGCGATAGTAGCCCCGAATAACGTTACCAAGACTGATACGCTGAAAGCAGATTCATATTTGGCCGCCTATTCAAAATCACGGATAGAAGCAGAAAAGGCAAAACAAGGCGTTATCATAGTGACCGCGAAAGACGGCGAAAAGAAAATATCGAATCGTGACGACAGTTCGTCTGGATATGATTATTCTCCACCGAGCGACAGTTTCAGTTCATCAGGATCGTCAAGTTTCTCCGGACCATCCAGTTACTCGCCATCTCCGAGTTACGGAACGCCAGTAAAATTCGAACCTGAAATTACCTATGACACTCCCCCTAATACTTATGGCCCTCCGTCCCCTAATTATGGTCCACCAAGCCATTCGTATGGACCTCCGGCACAAACATATGGCCCGCCGGCTCAGACATACGGACCACCGACTCAGTCGTATGGGCCACCGATTTACAAACCCATACTCCCAGTCTACGGACCGCCCCCGAAACCATCTTACGGTGTACCGTATGGCATGGCCGGTTTTAATTTCTTCGACAAACTGTCTTTGAAATTAGACATCCTAACGATTGCAAAGGTGCTCCTCAAATTTCTAATATTCAAGAAGATAGTGACAATGCTGGCTGTAGTTTGTATGTTGCTTGTGATACCAAAGCTAATCAATTTCAAAAAAGACGACGCTGTTAATAAGGATGAAGAGGACCGGAGCTATGGAGCTAGAAACGTGGTGCAGTTAACATCAGCGCAGCAGTTGTTGGAACGCGCAATCCTTGTCTACGGAAGGCAGGAACCTTCGTGTGGCGTTATTTGTCGAATGCGCAGAGTCGTCGACGACATTTACGAATTCCAACCTTATACTAGAAGTAACAATGTTGGCTCGTTGTTGTCATTATAA
Protein
MNSLQELQAIIKSKDNFTKFVINSEVPAVCKNEPCMLGVDEAGRGPVLGPMVYGVSYCPLNRKSILEELGCADSKALTEAKRDEIFTKMLTEEDSLNNVGWAVEVISPNYISNSMYRRAKCSLNEVSMNSAIDLIKKAAESGVNITEVYVDTVGPPEKYQTKLSEIFPSYKITVAKKADSIYPIVSAASIVAKVTRDHALKVWHFNEGLQLDHTEFGSGYPGDPLTKKFIRDRIDNVFGYPLLVRFSWSTAELMLQEKAAACTFEDVDDPGKGKIAIDGDNFLRAVAKFTIGKMLFRYCAALLALAVCASAIVAPNNVTKTDTLKADSYLAAYSKSRIEAEKAKQGVIIVTAKDGEKKISNRDDSSSGYDYSPPSDSFSSSGSSSFSGPSSYSPSPSYGTPVKFEPEITYDTPPNTYGPPSPNYGPPSHSYGPPAQTYGPPAQTYGPPTQSYGPPIYKPILPVYGPPPKPSYGVPYGMAGFNFFDKLSLKLDILTIAKVLLKFLIFKKIVTMLAVVCMLLVIPKLINFKKDDAVNKDEEDRSYGARNVVQLTSAQQLLERAILVYGRQEPSCGVICRMRRVVDDIYEFQPYTRSNNVGSLLSL
Summary
Description
Endonuclease that specifically degrades the RNA of RNA-DNA hybrids.
Catalytic Activity
Endonucleolytic cleavage to 5'-phosphomonoester.
Similarity
Belongs to the RNase HII family.
Uniprot
EC Number
3.1.26.4
Pubmed
Proteomes
PRIDE
Pfam
PF01351 RNase_HII
Interpro
SUPFAM
SSF53098
SSF53098
Gene 3D
ProteinModelPortal
PDB
3P5J
E-value=2.93069e-75,
Score=719
Ontologies
GO
PANTHER
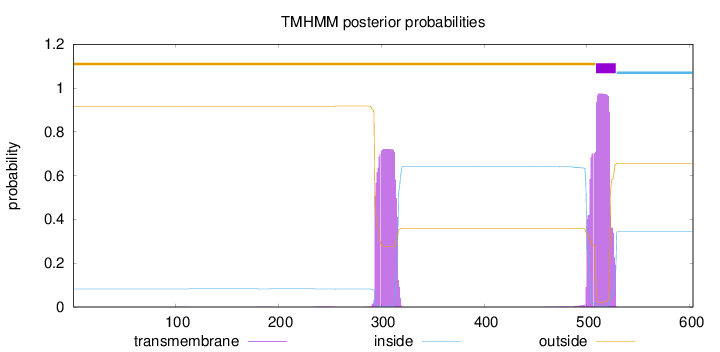
Topology
Length:
603
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
36.53703
Exp number, first 60 AAs:
0.00162
Total prob of N-in:
0.08269
outside
1 - 508
TMhelix
509 - 528
inside
529 - 603
Population Genetic Test Statistics
Pi
164.022316
Theta
174.217825
Tajima's D
-0.888147
CLR
4.469949
CSRT
0.155442227888606
Interpretation
Uncertain
