Gene
KWMTBOMO13763 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011531
Annotation
PREDICTED:_uncharacterized_protein_LOC106136946_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 2.202
Sequence
CDS
ATGGCCGTGTCGTTATACGTCGTGCTAGTGCTCGCCACGACAGCGCTAGGAGCGTCAATTGCGCCTTCAGAAACACAAGTCGTGGAAAAGTCATCATGTGGACGAGTTTGGGTGACAGTCCACTCTCCTGAGAAACGGTACTTGGGACAAACCTTGATTGATGTCTTGGAGCTTAATTGGGACTTCAGAGAATGCTCCACCATTCCTAGACAAATAGGTTTATTTGCCAATCGTCCCTTATCTTGGAACAATGCGATTGCAATTTTCGCCATCGGGTCCCCAGATGGTCACATAATCACTAACGTGTCCCTAGGAGAGGGCACGTTACCAGCAGGATGGGATACGGGAGCTGGGCCCAGAGGGCCTATCTGCTTATGGCCCTGGGTTGGAGCTGGAGACAATAACATAGAAGCCTTTAATTGTTTGAAGATACAGCCTACTTGGATGGAAGACAATGCAGATAAAATTAACAAACTTCGCATCGGTGAGTTAGCTCTGGCCGGCAGCCACAACGCGGGAGCGTGGCGCTTCGACACGGAACTCAGCAGCTTAGCCAGAGACAATTTCATTTTGTGCCAGGACCGCTCCATTTGGGCACAACTAGTCCACGGAATCAGATACTTAGACTTTAGGATTGGCTATTATGATTTTTACGACAAAGAGGAAGACAGATATTGGCTCAATCATAACTTAGTCAGGGTTCGACCTCTGGTACCGCTTTTGAGAGAAATCAGAGCGTTCTTGGACATCACAAATGAGCTGGTGTTCCTTGATGCCCATCACTTTCCAATCGGCTTCTACAATCCTGACGGAACACCGATCAGGGAAGTCCATCAAGGCTTGCTAAGAATTATACAAAGGGAGCTGGGACCGCACATAGCACCGGCCAATCGATTTGCCACCGGAGCCGGAAGCAGAGGTCCGACGGTACAGAGTCTGATTGATGCAAATCAAAGACTACACTTCAGTTACGTCGATAACATAATAGTTAATGAAAACAATTGGCTGTGGCCGAATCTGCCACATCTTTGGGCGAGAACTAACAATCCCACAGAGCTATTCCTATACTTAGACCAGGCGATTAGCGCGTCACCACAGCCGACGGCACGCTCGCCAATGCACTCAGCAATGGCCCAAACCACGCCTTCTGTTCTCGACATTCTATTGTTACGCGGCTCTCTGCGTATTAATGCTGATGGAGTTAATCGTAGGGTGACGGCCCGTCTCACATCACTCTGGCGACCAAACGCGAACATTGTCTCCACAGATTTCTTCCTCGGCAACGACGTCATCGACGTATCCATAGCGATTAGCGTGGAGCGTGCTGAGCAGTTATGA
Protein
MAVSLYVVLVLATTALGASIAPSETQVVEKSSCGRVWVTVHSPEKRYLGQTLIDVLELNWDFRECSTIPRQIGLFANRPLSWNNAIAIFAIGSPDGHIITNVSLGEGTLPAGWDTGAGPRGPICLWPWVGAGDNNIEAFNCLKIQPTWMEDNADKINKLRIGELALAGSHNAGAWRFDTELSSLARDNFILCQDRSIWAQLVHGIRYLDFRIGYYDFYDKEEDRYWLNHNLVRVRPLVPLLREIRAFLDITNELVFLDAHHFPIGFYNPDGTPIREVHQGLLRIIQRELGPHIAPANRFATGAGSRGPTVQSLIDANQRLHFSYVDNIIVNENNWLWPNLPHLWARTNNPTELFLYLDQAISASPQPTARSPMHSAMAQTTPSVLDILLLRGSLRINADGVNRRVTARLTSLWRPNANIVSTDFFLGNDVIDVSIAISVERAEQL
Summary
Uniprot
H9JPS6
A0A1E1W604
A0A2W1BGC5
A0A2H1WH39
A0A3S2NGZ5
A0A2A4J5M4
+ More
A0A0N1I3L7 A0A212EQ75 A0A194RUK9 A0A1E1W9D4 F4WWS0 A0A195DUJ2 A0A158NJ01 A0A0J7L0J2 A0A195D3G8 A0A195BBU0 E2BH78 A0A151X756 A0A0L7R927 A0A195FIB8 A0A026WFF2 A0A088ACV2 A0A088AT81 A0A154NWZ0 A0A2A3ET67 B7S909 A0A310SNX5 B7S8Q3 E2ARJ6 E9IIX7 A0A2J7QQG6 A0A2R7WEA1 A0A2J7QQF9 T1HBM1 A0A139WNB2 A0A1B6LSQ0 A0A1B6EUL8 A0A1Y1M5P8 A0A0K8SHX9 A0A0A9VXV5 A0A1Y1M9N4 N6TRU9 T1JAK6 E9FQM7 A0A0N8E6F2 A0A0P5ISK8 A0A0P5HLW0 A0A0P5P6C7 A0A0P5DP63 A0A0P5EKY8 A0A0N8A732 A0A0P5HPN4 A0A0P5AUG6 A0A0P6GW01 A0A0P5RLM9 A0A0P5LZP4 A0A0N8BSG0 A0A2L2XWT6 A0A2L2XXS3 A0A2L2Y089 A0A1B6CHD1 A0A087TY47 A0A0P5MA04 A0A0K8SJD1 A0A0A9VZI4 A0A1W4XF43 A0A0P5DYT5 A0A0K2TPA1 A0A0P5XXZ6 A0A2J7Q8Y8 A0A2J7Q8Y7 A0A0P6F6X5 A0A0P5CU89 A0A2K8JWY1 A0A139WNA6 T1J6B6 A0A067RX39 A0A1D2MY88 A0A2L2XVX3 T1KK76 A0A3R7PG65 A0A3S3QUH0 A0A2P6KJP6 A0A3S3NMK2 T1JAD6
A0A0N1I3L7 A0A212EQ75 A0A194RUK9 A0A1E1W9D4 F4WWS0 A0A195DUJ2 A0A158NJ01 A0A0J7L0J2 A0A195D3G8 A0A195BBU0 E2BH78 A0A151X756 A0A0L7R927 A0A195FIB8 A0A026WFF2 A0A088ACV2 A0A088AT81 A0A154NWZ0 A0A2A3ET67 B7S909 A0A310SNX5 B7S8Q3 E2ARJ6 E9IIX7 A0A2J7QQG6 A0A2R7WEA1 A0A2J7QQF9 T1HBM1 A0A139WNB2 A0A1B6LSQ0 A0A1B6EUL8 A0A1Y1M5P8 A0A0K8SHX9 A0A0A9VXV5 A0A1Y1M9N4 N6TRU9 T1JAK6 E9FQM7 A0A0N8E6F2 A0A0P5ISK8 A0A0P5HLW0 A0A0P5P6C7 A0A0P5DP63 A0A0P5EKY8 A0A0N8A732 A0A0P5HPN4 A0A0P5AUG6 A0A0P6GW01 A0A0P5RLM9 A0A0P5LZP4 A0A0N8BSG0 A0A2L2XWT6 A0A2L2XXS3 A0A2L2Y089 A0A1B6CHD1 A0A087TY47 A0A0P5MA04 A0A0K8SJD1 A0A0A9VZI4 A0A1W4XF43 A0A0P5DYT5 A0A0K2TPA1 A0A0P5XXZ6 A0A2J7Q8Y8 A0A2J7Q8Y7 A0A0P6F6X5 A0A0P5CU89 A0A2K8JWY1 A0A139WNA6 T1J6B6 A0A067RX39 A0A1D2MY88 A0A2L2XVX3 T1KK76 A0A3R7PG65 A0A3S3QUH0 A0A2P6KJP6 A0A3S3NMK2 T1JAD6
Pubmed
EMBL
BABH01015235
BABH01015236
BABH01015237
BABH01015238
BABH01015239
GDQN01008642
+ More
JAT82412.1 KZ150160 PZC72734.1 ODYU01008631 SOQ52378.1 RSAL01000042 RVE50830.1 NWSH01002927 PCG67271.1 KQ459037 KPJ04260.1 AGBW02013286 OWR43650.1 KQ459833 KPJ19826.1 GDQN01007470 JAT83584.1 GL888412 EGI61392.1 KQ980334 KYN16516.1 ADTU01017284 ADTU01017285 LBMM01001529 KMQ96136.1 KQ976885 KYN07442.1 KQ976530 KYM81664.1 GL448268 EFN84999.1 KQ982454 KYQ56159.1 KQ414630 KOC67296.1 KQ981523 KYN40148.1 KK107250 QOIP01000002 EZA54411.1 RLU25963.1 KQ434777 KZC04137.1 KZ288185 PBC34945.1 EF710656 ACE75384.1 KQ760672 OAD59456.1 EF710650 ACE75296.1 GL442102 EFN63944.1 GL763562 EFZ19458.1 NEVH01012087 PNF30829.1 KK854697 PTY17982.1 PNF30828.1 ACPB03008552 KQ971312 KYB29347.1 GEBQ01013281 JAT26696.1 GECZ01028168 JAS41601.1 GEZM01039973 JAV81044.1 GBRD01012930 GDHC01002208 JAG52896.1 JAQ16421.1 GBHO01042587 JAG01017.1 GEZM01039972 JAV81045.1 APGK01021776 KB740363 KB631964 ENN80803.1 ERL87504.1 JH432001 GL732523 EFX90339.1 GDIQ01057252 JAN37485.1 GDIQ01209647 JAK42078.1 GDIQ01226143 GDIP01012239 JAK25582.1 JAM91476.1 GDIQ01132983 JAL18743.1 GDIP01156895 JAJ66507.1 GDIP01158304 JAJ65098.1 GDIP01176081 JAJ47321.1 GDIP01237181 GDIP01235385 GDIP01220990 GDIP01211586 GDIP01184483 GDIP01183336 GDIP01176083 GDIP01163237 GDIQ01224989 GDIP01138095 GDIP01085546 LRGB01000642 JAI86220.1 JAK26736.1 KZS17294.1 GDIP01254269 GDIP01251128 GDIP01232483 GDIP01229927 GDIP01224776 GDIP01213529 GDIP01210678 GDIP01208133 GDIP01185730 GDIP01183335 GDIP01176082 GDIP01156041 GDIP01154599 GDIP01153136 GDIP01137589 GDIP01125192 GDIP01124196 GDIP01123039 GDIP01121489 GDIP01119598 GDIP01112844 GDIP01111792 GDIP01108141 GDIP01102799 GDIP01097010 GDIP01086793 GDIP01083948 GDIP01079894 GDIP01070500 GDIP01053267 GDIP01052099 GDIP01046354 GDIP01044897 GDIP01040402 JAJ15269.1 GDIQ01028411 JAN66326.1 GDIQ01117986 JAL33740.1 GDIQ01173512 JAK78213.1 GDIQ01149379 JAL02347.1 IAAA01006096 LAA00529.1 IAAA01006095 LAA00527.1 IAAA01006094 LAA00525.1 GEDC01024507 JAS12791.1 KK117282 KFM70036.1 GDIQ01167207 JAK84518.1 GBRD01012931 JAG52895.1 GBHO01042585 JAG01019.1 GDIP01148999 JAJ74403.1 HACA01010324 CDW27685.1 GDIP01065603 JAM38112.1 NEVH01016945 PNF25049.1 PNF25050.1 GDIQ01053770 JAN40967.1 GDIP01166434 JAJ56968.1 KY031176 ATU82927.1 KYB29346.1 JH431874 KK852411 KDR24464.1 LJIJ01000389 ODM98013.1 IAAA01002090 LAA00246.1 CAEY01000175 QCYY01000002 ROT86099.1 NCKU01000793 RWS14140.1 MWRG01010394 PRD26527.1 NCKU01007734 RWS02432.1 JH431989
JAT82412.1 KZ150160 PZC72734.1 ODYU01008631 SOQ52378.1 RSAL01000042 RVE50830.1 NWSH01002927 PCG67271.1 KQ459037 KPJ04260.1 AGBW02013286 OWR43650.1 KQ459833 KPJ19826.1 GDQN01007470 JAT83584.1 GL888412 EGI61392.1 KQ980334 KYN16516.1 ADTU01017284 ADTU01017285 LBMM01001529 KMQ96136.1 KQ976885 KYN07442.1 KQ976530 KYM81664.1 GL448268 EFN84999.1 KQ982454 KYQ56159.1 KQ414630 KOC67296.1 KQ981523 KYN40148.1 KK107250 QOIP01000002 EZA54411.1 RLU25963.1 KQ434777 KZC04137.1 KZ288185 PBC34945.1 EF710656 ACE75384.1 KQ760672 OAD59456.1 EF710650 ACE75296.1 GL442102 EFN63944.1 GL763562 EFZ19458.1 NEVH01012087 PNF30829.1 KK854697 PTY17982.1 PNF30828.1 ACPB03008552 KQ971312 KYB29347.1 GEBQ01013281 JAT26696.1 GECZ01028168 JAS41601.1 GEZM01039973 JAV81044.1 GBRD01012930 GDHC01002208 JAG52896.1 JAQ16421.1 GBHO01042587 JAG01017.1 GEZM01039972 JAV81045.1 APGK01021776 KB740363 KB631964 ENN80803.1 ERL87504.1 JH432001 GL732523 EFX90339.1 GDIQ01057252 JAN37485.1 GDIQ01209647 JAK42078.1 GDIQ01226143 GDIP01012239 JAK25582.1 JAM91476.1 GDIQ01132983 JAL18743.1 GDIP01156895 JAJ66507.1 GDIP01158304 JAJ65098.1 GDIP01176081 JAJ47321.1 GDIP01237181 GDIP01235385 GDIP01220990 GDIP01211586 GDIP01184483 GDIP01183336 GDIP01176083 GDIP01163237 GDIQ01224989 GDIP01138095 GDIP01085546 LRGB01000642 JAI86220.1 JAK26736.1 KZS17294.1 GDIP01254269 GDIP01251128 GDIP01232483 GDIP01229927 GDIP01224776 GDIP01213529 GDIP01210678 GDIP01208133 GDIP01185730 GDIP01183335 GDIP01176082 GDIP01156041 GDIP01154599 GDIP01153136 GDIP01137589 GDIP01125192 GDIP01124196 GDIP01123039 GDIP01121489 GDIP01119598 GDIP01112844 GDIP01111792 GDIP01108141 GDIP01102799 GDIP01097010 GDIP01086793 GDIP01083948 GDIP01079894 GDIP01070500 GDIP01053267 GDIP01052099 GDIP01046354 GDIP01044897 GDIP01040402 JAJ15269.1 GDIQ01028411 JAN66326.1 GDIQ01117986 JAL33740.1 GDIQ01173512 JAK78213.1 GDIQ01149379 JAL02347.1 IAAA01006096 LAA00529.1 IAAA01006095 LAA00527.1 IAAA01006094 LAA00525.1 GEDC01024507 JAS12791.1 KK117282 KFM70036.1 GDIQ01167207 JAK84518.1 GBRD01012931 JAG52895.1 GBHO01042585 JAG01019.1 GDIP01148999 JAJ74403.1 HACA01010324 CDW27685.1 GDIP01065603 JAM38112.1 NEVH01016945 PNF25049.1 PNF25050.1 GDIQ01053770 JAN40967.1 GDIP01166434 JAJ56968.1 KY031176 ATU82927.1 KYB29346.1 JH431874 KK852411 KDR24464.1 LJIJ01000389 ODM98013.1 IAAA01002090 LAA00246.1 CAEY01000175 QCYY01000002 ROT86099.1 NCKU01000793 RWS14140.1 MWRG01010394 PRD26527.1 NCKU01007734 RWS02432.1 JH431989
Proteomes
UP000005204
UP000283053
UP000218220
UP000053268
UP000007151
UP000053240
+ More
UP000007755 UP000078492 UP000005205 UP000036403 UP000078542 UP000078540 UP000008237 UP000075809 UP000053825 UP000078541 UP000053097 UP000279307 UP000005203 UP000076502 UP000242457 UP000000311 UP000235965 UP000015103 UP000007266 UP000019118 UP000030742 UP000000305 UP000076858 UP000054359 UP000192223 UP000027135 UP000094527 UP000015104 UP000283509 UP000285301
UP000007755 UP000078492 UP000005205 UP000036403 UP000078542 UP000078540 UP000008237 UP000075809 UP000053825 UP000078541 UP000053097 UP000279307 UP000005203 UP000076502 UP000242457 UP000000311 UP000235965 UP000015103 UP000007266 UP000019118 UP000030742 UP000000305 UP000076858 UP000054359 UP000192223 UP000027135 UP000094527 UP000015104 UP000283509 UP000285301
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JPS6
A0A1E1W604
A0A2W1BGC5
A0A2H1WH39
A0A3S2NGZ5
A0A2A4J5M4
+ More
A0A0N1I3L7 A0A212EQ75 A0A194RUK9 A0A1E1W9D4 F4WWS0 A0A195DUJ2 A0A158NJ01 A0A0J7L0J2 A0A195D3G8 A0A195BBU0 E2BH78 A0A151X756 A0A0L7R927 A0A195FIB8 A0A026WFF2 A0A088ACV2 A0A088AT81 A0A154NWZ0 A0A2A3ET67 B7S909 A0A310SNX5 B7S8Q3 E2ARJ6 E9IIX7 A0A2J7QQG6 A0A2R7WEA1 A0A2J7QQF9 T1HBM1 A0A139WNB2 A0A1B6LSQ0 A0A1B6EUL8 A0A1Y1M5P8 A0A0K8SHX9 A0A0A9VXV5 A0A1Y1M9N4 N6TRU9 T1JAK6 E9FQM7 A0A0N8E6F2 A0A0P5ISK8 A0A0P5HLW0 A0A0P5P6C7 A0A0P5DP63 A0A0P5EKY8 A0A0N8A732 A0A0P5HPN4 A0A0P5AUG6 A0A0P6GW01 A0A0P5RLM9 A0A0P5LZP4 A0A0N8BSG0 A0A2L2XWT6 A0A2L2XXS3 A0A2L2Y089 A0A1B6CHD1 A0A087TY47 A0A0P5MA04 A0A0K8SJD1 A0A0A9VZI4 A0A1W4XF43 A0A0P5DYT5 A0A0K2TPA1 A0A0P5XXZ6 A0A2J7Q8Y8 A0A2J7Q8Y7 A0A0P6F6X5 A0A0P5CU89 A0A2K8JWY1 A0A139WNA6 T1J6B6 A0A067RX39 A0A1D2MY88 A0A2L2XVX3 T1KK76 A0A3R7PG65 A0A3S3QUH0 A0A2P6KJP6 A0A3S3NMK2 T1JAD6
A0A0N1I3L7 A0A212EQ75 A0A194RUK9 A0A1E1W9D4 F4WWS0 A0A195DUJ2 A0A158NJ01 A0A0J7L0J2 A0A195D3G8 A0A195BBU0 E2BH78 A0A151X756 A0A0L7R927 A0A195FIB8 A0A026WFF2 A0A088ACV2 A0A088AT81 A0A154NWZ0 A0A2A3ET67 B7S909 A0A310SNX5 B7S8Q3 E2ARJ6 E9IIX7 A0A2J7QQG6 A0A2R7WEA1 A0A2J7QQF9 T1HBM1 A0A139WNB2 A0A1B6LSQ0 A0A1B6EUL8 A0A1Y1M5P8 A0A0K8SHX9 A0A0A9VXV5 A0A1Y1M9N4 N6TRU9 T1JAK6 E9FQM7 A0A0N8E6F2 A0A0P5ISK8 A0A0P5HLW0 A0A0P5P6C7 A0A0P5DP63 A0A0P5EKY8 A0A0N8A732 A0A0P5HPN4 A0A0P5AUG6 A0A0P6GW01 A0A0P5RLM9 A0A0P5LZP4 A0A0N8BSG0 A0A2L2XWT6 A0A2L2XXS3 A0A2L2Y089 A0A1B6CHD1 A0A087TY47 A0A0P5MA04 A0A0K8SJD1 A0A0A9VZI4 A0A1W4XF43 A0A0P5DYT5 A0A0K2TPA1 A0A0P5XXZ6 A0A2J7Q8Y8 A0A2J7Q8Y7 A0A0P6F6X5 A0A0P5CU89 A0A2K8JWY1 A0A139WNA6 T1J6B6 A0A067RX39 A0A1D2MY88 A0A2L2XVX3 T1KK76 A0A3R7PG65 A0A3S3QUH0 A0A2P6KJP6 A0A3S3NMK2 T1JAD6
Ontologies
GO
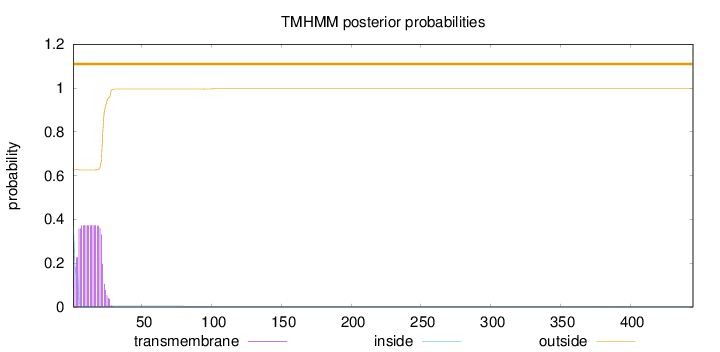
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.992065
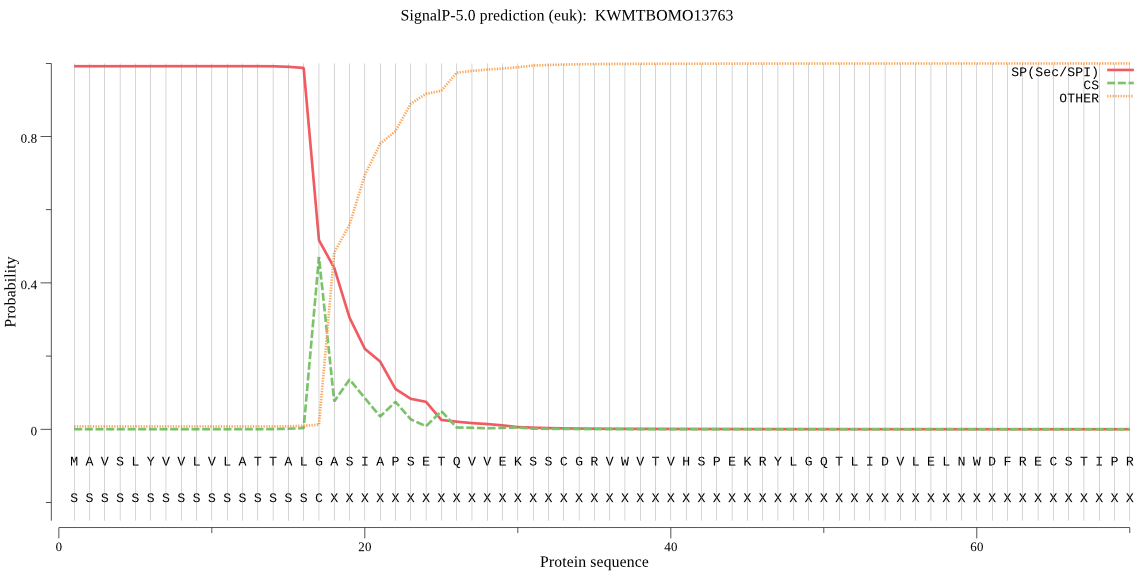
Length:
445
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.45545999999999
Exp number, first 60 AAs:
7.40764
Total prob of N-in:
0.37097
outside
1 - 445
Population Genetic Test Statistics
Pi
30.203216
Theta
172.742413
Tajima's D
1.113143
CLR
0.384215
CSRT
0.690565471726414
Interpretation
Uncertain
