Gene
KWMTBOMO13762
Pre Gene Modal
BGIBMGA011530
Annotation
ionotropic_glutamate_receptor_[Helicoverpa_armigera]
Location in the cell
Cytoplasmic Reliability : 1.632 Nuclear Reliability : 1.089 PlasmaMembrane Reliability : 1.325
Sequence
CDS
ATGCTGCAGCCTCCCAGAACTGACAGCTACTCAGTTTGGAGATCGTTATGCTCCAATACTGCAATGAGGCCAGTTGCAGTTTTTGGACCTCAAAACCCAATCTCCGACAAGGCAGTTCGTGATCAATGCGCTATTGCTAACATACCTCACATACAAGCCACAATGAAACCACTAGATCCAGATCTTGAACTTAATGAGGAATTAGATTCAAGTAATTCCCAAATTGAAACCGCCGATGAGGCTGCTGCTTTTAAGAAAATAACCATAAATTTTTACCCAGAATCAGACGAAATATCTGTAGCTTACGGGAAACTACTAAATTTTTACAACTGGAAGGTTTTCGCAGTTCTTTACGAAGACGATTTTGGATTACTTCGCATTCAAAAGATACTCGGCAGACATAGTGACGACTATCCAGTAACTGTGTATAGATTAGATCCAGAGGGAGACAACAGAAGTATATTCAAACTTCTGAATGAAGACAGAGAAGATCGAATTTTATTGGATTGTCATGTAGACAGAGTTATGAAATATATGGAGGAAGCTGGTAGCTTCAAGAACTTGAGCAACTATCAGCACTACATCTTAGTAACCTTGGATGCAGCTACGATTTATGATAAATTAATAAACCTTCCTTTAAATGTAACCTGGTTAAGCCTGACAGAATTTGAAAAAATTAAAGATGCTCAACATTATTTAGCGACTAGAGTTGGAAAATGGTCTAATGACTTTTCAACCAAAGTCAATCAGATGAAGACTGATGAATTGGTAATGGATGATATTACCAACCACGTGGTGAAAGCAATGGAAACAATAGAAGACTTAAGAAAACCAGATTTTCCAATATGCGAAAAAGATCCACATCCCTGGCCACATGGAGCACAACTACAAAAAGCTATACTGCAGACTAAAACTGTCGGTGTTACTGGTAACATAGAATTCAACGAGAACGGCAAAAGGATAAACTACGTTTTGTTTGTAAATGAAATTTACATGTCGCAAAGATATACTATAGGGAAATGGGATTCATCCAATGCCACTGAAATTGTCGAAACCAGACAAACAGATAATCGCACTACTGCGCAACAAAGTAAACATTTTATTGTAATTTCTAGAAGAAACAAACCGTATTTTTACGAGAAGGAACCATGCAAGGAGGGAGACACCACATGTGTGAACGATAAAGAGAAATACGAAGGATTCTCTGTTGATCTCGTAAAGGCCATTTTCGAGATTTTAAAAAGCGAAAACTTTAATTACACTTACTCCTTCTTCCATGACGAGGACAAAGATTATGGGGAATATGACGTGAAAACAAAGAAATGGAATGGCTTAATCGGTGACTTACTCGATAAGAAAGCAGACTTAGCCGTCTGTGATCTAACAATCACTGAAGATCGAAAGAAGATTGTGGATTTTTCAGTTCCATTCATGTCGTTAGGCATAAGTATATTGTTTAAGAAACAGCTTGCGGAACCACCTGGAATGTTTTCTTTCCTCAATCCATATAGTGTCGGGGTATGGCTGTACACAGCTACCGCTTATTGCGTCGTTTCAATGGTGCTATTTGTTTGTTCCAGAATTTCACCTGCTGATTGGGAAAACCCGGAACCTTGCGAGAAAAATCCAGAAGAATTAGAAAATATTTGGACATTCAAGAACTGTGCGTGGTTGACAATGGGATCTATCATGACACAAGGATGTGATATCTTACCCAAGGCTATCGGAACCCGTTGGGTCTGCGGAATGTGGTGGTTCTTCTCTATGATTGTGTGTCAAACATATATCGCCCAGCTGTCGGCTTCGATGACAACCGCTTTGGAAAGTGAATCTATTAAAAGCGTAGAAGATTTAGCGGCTCAAAACAAAATACTATACGGCGCTATTAAGGGGGGATCTACAGTTGATTTTTTTAAGAACTCCAAAGACAAAACCTTCAGACGTATGTATGAAACAATGAACTCAAATCCAGCATTGCTAGTCAAAAGTAACGATGAAGGAGTGAGAAGGGTTTTAAGCGGATCTAACAAATACGCTTTCTTCATGGAATCTTCTGCCATTGAATACAAGCTTAGAAGAGACTGTAGACTGAAAAAAATAGGAGGGGAACTGGACAACAAATACTATGGGATCGCTATGCCAGCAAATTCTCCTTTTCGAACTCATATAAATAGGGCTATATTGAAACTACAAGAGTCGACAAAGTTAGATAAAATCAGGAACTTATGGTGGTTACAAAAGTATGATGCGAAAATCTGCAAGGATGTAGACGTTGACGCGAACGACGTACAAGGGGACTTGGAGCTTGAAAATCTTCTAGGTGCTTTCGTTGTACTGATAGTTGGCATATTCTTTAGCCTCATTATTACCGCCATAGAATTCATGAACGAAGTCAGGAATATCGTTGTTCGTGAAAATGTCACACACAAAGAAGTTCTTGTCAAAGAATTAAAAGCTTCGCTGAACTTTTTCAACAAGCAGAAGCCAGTACTGAGGAACCCGAGCCGTGCTCCATCGTTGGCAGGCACAGAGGAATCTGAAGATAAAAACAAAGAGAAGAAGCAGAAGAAAGAAAAAAGCATTGAAAACTTTATGGATTTCGAAAAAGGAGCTTCGATGAATAATTAA
Protein
MLQPPRTDSYSVWRSLCSNTAMRPVAVFGPQNPISDKAVRDQCAIANIPHIQATMKPLDPDLELNEELDSSNSQIETADEAAAFKKITINFYPESDEISVAYGKLLNFYNWKVFAVLYEDDFGLLRIQKILGRHSDDYPVTVYRLDPEGDNRSIFKLLNEDREDRILLDCHVDRVMKYMEEAGSFKNLSNYQHYILVTLDAATIYDKLINLPLNVTWLSLTEFEKIKDAQHYLATRVGKWSNDFSTKVNQMKTDELVMDDITNHVVKAMETIEDLRKPDFPICEKDPHPWPHGAQLQKAILQTKTVGVTGNIEFNENGKRINYVLFVNEIYMSQRYTIGKWDSSNATEIVETRQTDNRTTAQQSKHFIVISRRNKPYFYEKEPCKEGDTTCVNDKEKYEGFSVDLVKAIFEILKSENFNYTYSFFHDEDKDYGEYDVKTKKWNGLIGDLLDKKADLAVCDLTITEDRKKIVDFSVPFMSLGISILFKKQLAEPPGMFSFLNPYSVGVWLYTATAYCVVSMVLFVCSRISPADWENPEPCEKNPEELENIWTFKNCAWLTMGSIMTQGCDILPKAIGTRWVCGMWWFFSMIVCQTYIAQLSASMTTALESESIKSVEDLAAQNKILYGAIKGGSTVDFFKNSKDKTFRRMYETMNSNPALLVKSNDEGVRRVLSGSNKYAFFMESSAIEYKLRRDCRLKKIGGELDNKYYGIAMPANSPFRTHINRAILKLQESTKLDKIRNLWWLQKYDAKICKDVDVDANDVQGDLELENLLGAFVVLIVGIFFSLIITAIEFMNEVRNIVVRENVTHKEVLVKELKASLNFFNKQKPVLRNPSRAPSLAGTEESEDKNKEKKQKKEKSIENFMDFEKGASMNN
Summary
Similarity
Belongs to the glutamate-gated ion channel (TC 1.A.10.1) family.
Uniprot
A0A0B5A1Y6
A0A2H1WCB3
A0A075T678
A0A0N1PH66
A0A212EQS2
A0A194RR91
+ More
A0A2H1W6P8 A0A2W1BMM8 A0A2A4J734 A0A212EQW1 A0A182H4S6 A0A0N1PG13 Q17HY6 A0A1S4F1W5 A0A3S2NWQ7 A0A1Q3FZA3 A0A182H4S5 A0A182GWX5 A0A1Q3FZ95 Q7QHV3 A0A084WPK0 A0A182XPJ9 A0A182HFV7 A0A182JKW8 Q17HY5 A0A1S4F219 A0A182VIJ3 A0A3S2M3Z8 A0A182TWK6 A0A182N102 A0A194RPP3 W5JDY5 A0A1L8DK79 A0A182VMN8 A0A182P835 Q7QE65 A0A182LIW7 A0A182FK06 A0A182M854 A0A182KFG5 T1H819 A0A182Q917 A0A182RJ06 A0A182I9N5 A0A182YEZ2 A0A182X5A0 A0A1Y9GKF3 A0A2J7QQG3 Q7PRP7 A0A195DUH1 A0A1S4G9Z3 A0A158NJ03 A0A195FIP6 A0A0M4JB15 A0A2M4BDE6 F5HJZ1 A0A084WPK1 A0A1W4XE85 A0A1S4GA38 A0A2J7QQK9 A0A1W4XFF8 A0A336KHH4 A0A1W4XF50 A0A2J7QQG1 E0VK11 T1PMA4 A0A3L8DZQ4 A0A2U9NJD5 A0A0A1XPV5 A0A140CQC2 A0A1I8NU50 A0A1S4EBS0 A0A2J7QQL5 A0A1W4X4N7 A0A2J7QQL0 A0A0K8V3R8 A0A034V5K6 A0A067QWX4 A0A1I8MEH0 A0A2J7QQG8 A0A1S4EC64 A0A2H4ZBE6 A0A223HCX9 A0A345BF16 A0A0V0J2K9 A0A0F7QEG0 H9JPS3 A0A023F5M1 A0A075T818 A0A2A4J849 B4IFM4 A0A0J9R5B5 A0A139WN68 A0A3S2NLN9 D6X0W1 A0A0J9R4W9 A0A0J9R6E4 B3NKS8
A0A2H1W6P8 A0A2W1BMM8 A0A2A4J734 A0A212EQW1 A0A182H4S6 A0A0N1PG13 Q17HY6 A0A1S4F1W5 A0A3S2NWQ7 A0A1Q3FZA3 A0A182H4S5 A0A182GWX5 A0A1Q3FZ95 Q7QHV3 A0A084WPK0 A0A182XPJ9 A0A182HFV7 A0A182JKW8 Q17HY5 A0A1S4F219 A0A182VIJ3 A0A3S2M3Z8 A0A182TWK6 A0A182N102 A0A194RPP3 W5JDY5 A0A1L8DK79 A0A182VMN8 A0A182P835 Q7QE65 A0A182LIW7 A0A182FK06 A0A182M854 A0A182KFG5 T1H819 A0A182Q917 A0A182RJ06 A0A182I9N5 A0A182YEZ2 A0A182X5A0 A0A1Y9GKF3 A0A2J7QQG3 Q7PRP7 A0A195DUH1 A0A1S4G9Z3 A0A158NJ03 A0A195FIP6 A0A0M4JB15 A0A2M4BDE6 F5HJZ1 A0A084WPK1 A0A1W4XE85 A0A1S4GA38 A0A2J7QQK9 A0A1W4XFF8 A0A336KHH4 A0A1W4XF50 A0A2J7QQG1 E0VK11 T1PMA4 A0A3L8DZQ4 A0A2U9NJD5 A0A0A1XPV5 A0A140CQC2 A0A1I8NU50 A0A1S4EBS0 A0A2J7QQL5 A0A1W4X4N7 A0A2J7QQL0 A0A0K8V3R8 A0A034V5K6 A0A067QWX4 A0A1I8MEH0 A0A2J7QQG8 A0A1S4EC64 A0A2H4ZBE6 A0A223HCX9 A0A345BF16 A0A0V0J2K9 A0A0F7QEG0 H9JPS3 A0A023F5M1 A0A075T818 A0A2A4J849 B4IFM4 A0A0J9R5B5 A0A139WN68 A0A3S2NLN9 D6X0W1 A0A0J9R4W9 A0A0J9R6E4 B3NKS8
Pubmed
25027790
26354079
22118469
28756777
26483478
17510324
+ More
12364791 14747013 17210077 24438588 20920257 23761445 20966253 25244985 21347285 26265180 20566863 30249741 25830018 26752702 25348373 24845553 25315136 29133804 28150741 29727827 27006164 25803580 19121390 25474469 17994087 22936249 18362917 19820115
12364791 14747013 17210077 24438588 20920257 23761445 20966253 25244985 21347285 26265180 20566863 30249741 25830018 26752702 25348373 24845553 25315136 29133804 28150741 29727827 27006164 25803580 19121390 25474469 17994087 22936249 18362917 19820115
EMBL
KJ542734
AJD81618.1
ODYU01007642
SOQ50596.1
KF768736
AIG51930.1
+ More
KQ459037 KPJ04259.1 AGBW02013238 OWR43843.1 KQ459833 KPJ19825.1 ODYU01006689 SOQ48759.1 KZ150075 PZC73970.1 NWSH01002576 PCG67937.1 OWR43841.1 JXUM01110194 KQ565389 KXJ71034.1 KPJ04257.1 CH477244 EAT46290.1 RSAL01000042 RVE50833.1 GFDL01002185 JAV32860.1 KXJ71033.1 JXUM01094236 KQ564114 KXJ72756.1 GFDL01002179 JAV32866.1 AAAB01008815 EAA05023.4 ATLV01025084 KE525369 KFB52144.1 APCN01007369 EAT46291.1 RVE50832.1 KPJ19823.1 ADMH02001821 ETN61009.1 GFDF01007263 JAV06821.1 AAAB01008847 EAA06913.4 AXCM01004371 ACPB03008552 AXCN02001376 APCN01001012 APCN01001013 APCN01001007 APCN01001008 APCN01001009 APCN01001010 APCN01001011 NEVH01012087 PNF30825.1 EAA06836.6 KQ980334 KYN16523.1 ADTU01017309 ADTU01017310 ADTU01017311 ADTU01017312 ADTU01017313 ADTU01017314 ADTU01017315 ADTU01017316 ADTU01017317 ADTU01017318 KQ981523 KYN40142.1 KP843209 ALD51346.1 GGFJ01001924 MBW51065.1 EGK96607.1 ATLV01025088 ATLV01025089 ATLV01025090 ATLV01025091 ATLV01025092 KFB52145.1 PNF30870.1 UFQS01000433 UFQT01000433 SSX03889.1 SSX24254.1 PNF30824.1 DS235235 EEB13717.1 KA649912 AFP64541.1 QOIP01000002 RLU25960.1 MF385087 AWT23259.1 GBXI01001226 JAD13066.1 KR998364 AMH85983.1 PNF30867.1 PNF30869.1 GDHF01018808 GDHF01006355 JAI33506.1 JAI45959.1 GAKP01021897 JAC37055.1 KK853270 KDR09166.1 PNF30826.1 MF975501 AUF73077.1 KY283580 AST36240.1 MG820669 AXF48840.1 GDKB01000032 JAP38464.1 LC017781 BAR64795.1 BABH01015229 GBBI01002254 JAC16458.1 KF768733 AIG51927.1 PCG67936.1 CH480833 EDW46446.1 CM002910 KMY91308.1 KQ971312 KYB29344.1 RVE50836.1 KQ971372 EFA10562.2 KMY91307.1 KMY91304.1 CH954179 EDV54451.2
KQ459037 KPJ04259.1 AGBW02013238 OWR43843.1 KQ459833 KPJ19825.1 ODYU01006689 SOQ48759.1 KZ150075 PZC73970.1 NWSH01002576 PCG67937.1 OWR43841.1 JXUM01110194 KQ565389 KXJ71034.1 KPJ04257.1 CH477244 EAT46290.1 RSAL01000042 RVE50833.1 GFDL01002185 JAV32860.1 KXJ71033.1 JXUM01094236 KQ564114 KXJ72756.1 GFDL01002179 JAV32866.1 AAAB01008815 EAA05023.4 ATLV01025084 KE525369 KFB52144.1 APCN01007369 EAT46291.1 RVE50832.1 KPJ19823.1 ADMH02001821 ETN61009.1 GFDF01007263 JAV06821.1 AAAB01008847 EAA06913.4 AXCM01004371 ACPB03008552 AXCN02001376 APCN01001012 APCN01001013 APCN01001007 APCN01001008 APCN01001009 APCN01001010 APCN01001011 NEVH01012087 PNF30825.1 EAA06836.6 KQ980334 KYN16523.1 ADTU01017309 ADTU01017310 ADTU01017311 ADTU01017312 ADTU01017313 ADTU01017314 ADTU01017315 ADTU01017316 ADTU01017317 ADTU01017318 KQ981523 KYN40142.1 KP843209 ALD51346.1 GGFJ01001924 MBW51065.1 EGK96607.1 ATLV01025088 ATLV01025089 ATLV01025090 ATLV01025091 ATLV01025092 KFB52145.1 PNF30870.1 UFQS01000433 UFQT01000433 SSX03889.1 SSX24254.1 PNF30824.1 DS235235 EEB13717.1 KA649912 AFP64541.1 QOIP01000002 RLU25960.1 MF385087 AWT23259.1 GBXI01001226 JAD13066.1 KR998364 AMH85983.1 PNF30867.1 PNF30869.1 GDHF01018808 GDHF01006355 JAI33506.1 JAI45959.1 GAKP01021897 JAC37055.1 KK853270 KDR09166.1 PNF30826.1 MF975501 AUF73077.1 KY283580 AST36240.1 MG820669 AXF48840.1 GDKB01000032 JAP38464.1 LC017781 BAR64795.1 BABH01015229 GBBI01002254 JAC16458.1 KF768733 AIG51927.1 PCG67936.1 CH480833 EDW46446.1 CM002910 KMY91308.1 KQ971312 KYB29344.1 RVE50836.1 KQ971372 EFA10562.2 KMY91307.1 KMY91304.1 CH954179 EDV54451.2
Proteomes
UP000053268
UP000007151
UP000053240
UP000218220
UP000069940
UP000249989
+ More
UP000008820 UP000283053 UP000007062 UP000030765 UP000076407 UP000075840 UP000075880 UP000075903 UP000075902 UP000075884 UP000000673 UP000075885 UP000075882 UP000069272 UP000075883 UP000075881 UP000015103 UP000075886 UP000075900 UP000076408 UP000235965 UP000078492 UP000005205 UP000078541 UP000192223 UP000009046 UP000279307 UP000095300 UP000079169 UP000027135 UP000095301 UP000005204 UP000001292 UP000007266 UP000008711
UP000008820 UP000283053 UP000007062 UP000030765 UP000076407 UP000075840 UP000075880 UP000075903 UP000075902 UP000075884 UP000000673 UP000075885 UP000075882 UP000069272 UP000075883 UP000075881 UP000015103 UP000075886 UP000075900 UP000076408 UP000235965 UP000078492 UP000005205 UP000078541 UP000192223 UP000009046 UP000279307 UP000095300 UP000079169 UP000027135 UP000095301 UP000005204 UP000001292 UP000007266 UP000008711
PRIDE
Interpro
SUPFAM
SSF53822
SSF53822
ProteinModelPortal
A0A0B5A1Y6
A0A2H1WCB3
A0A075T678
A0A0N1PH66
A0A212EQS2
A0A194RR91
+ More
A0A2H1W6P8 A0A2W1BMM8 A0A2A4J734 A0A212EQW1 A0A182H4S6 A0A0N1PG13 Q17HY6 A0A1S4F1W5 A0A3S2NWQ7 A0A1Q3FZA3 A0A182H4S5 A0A182GWX5 A0A1Q3FZ95 Q7QHV3 A0A084WPK0 A0A182XPJ9 A0A182HFV7 A0A182JKW8 Q17HY5 A0A1S4F219 A0A182VIJ3 A0A3S2M3Z8 A0A182TWK6 A0A182N102 A0A194RPP3 W5JDY5 A0A1L8DK79 A0A182VMN8 A0A182P835 Q7QE65 A0A182LIW7 A0A182FK06 A0A182M854 A0A182KFG5 T1H819 A0A182Q917 A0A182RJ06 A0A182I9N5 A0A182YEZ2 A0A182X5A0 A0A1Y9GKF3 A0A2J7QQG3 Q7PRP7 A0A195DUH1 A0A1S4G9Z3 A0A158NJ03 A0A195FIP6 A0A0M4JB15 A0A2M4BDE6 F5HJZ1 A0A084WPK1 A0A1W4XE85 A0A1S4GA38 A0A2J7QQK9 A0A1W4XFF8 A0A336KHH4 A0A1W4XF50 A0A2J7QQG1 E0VK11 T1PMA4 A0A3L8DZQ4 A0A2U9NJD5 A0A0A1XPV5 A0A140CQC2 A0A1I8NU50 A0A1S4EBS0 A0A2J7QQL5 A0A1W4X4N7 A0A2J7QQL0 A0A0K8V3R8 A0A034V5K6 A0A067QWX4 A0A1I8MEH0 A0A2J7QQG8 A0A1S4EC64 A0A2H4ZBE6 A0A223HCX9 A0A345BF16 A0A0V0J2K9 A0A0F7QEG0 H9JPS3 A0A023F5M1 A0A075T818 A0A2A4J849 B4IFM4 A0A0J9R5B5 A0A139WN68 A0A3S2NLN9 D6X0W1 A0A0J9R4W9 A0A0J9R6E4 B3NKS8
A0A2H1W6P8 A0A2W1BMM8 A0A2A4J734 A0A212EQW1 A0A182H4S6 A0A0N1PG13 Q17HY6 A0A1S4F1W5 A0A3S2NWQ7 A0A1Q3FZA3 A0A182H4S5 A0A182GWX5 A0A1Q3FZ95 Q7QHV3 A0A084WPK0 A0A182XPJ9 A0A182HFV7 A0A182JKW8 Q17HY5 A0A1S4F219 A0A182VIJ3 A0A3S2M3Z8 A0A182TWK6 A0A182N102 A0A194RPP3 W5JDY5 A0A1L8DK79 A0A182VMN8 A0A182P835 Q7QE65 A0A182LIW7 A0A182FK06 A0A182M854 A0A182KFG5 T1H819 A0A182Q917 A0A182RJ06 A0A182I9N5 A0A182YEZ2 A0A182X5A0 A0A1Y9GKF3 A0A2J7QQG3 Q7PRP7 A0A195DUH1 A0A1S4G9Z3 A0A158NJ03 A0A195FIP6 A0A0M4JB15 A0A2M4BDE6 F5HJZ1 A0A084WPK1 A0A1W4XE85 A0A1S4GA38 A0A2J7QQK9 A0A1W4XFF8 A0A336KHH4 A0A1W4XF50 A0A2J7QQG1 E0VK11 T1PMA4 A0A3L8DZQ4 A0A2U9NJD5 A0A0A1XPV5 A0A140CQC2 A0A1I8NU50 A0A1S4EBS0 A0A2J7QQL5 A0A1W4X4N7 A0A2J7QQL0 A0A0K8V3R8 A0A034V5K6 A0A067QWX4 A0A1I8MEH0 A0A2J7QQG8 A0A1S4EC64 A0A2H4ZBE6 A0A223HCX9 A0A345BF16 A0A0V0J2K9 A0A0F7QEG0 H9JPS3 A0A023F5M1 A0A075T818 A0A2A4J849 B4IFM4 A0A0J9R5B5 A0A139WN68 A0A3S2NLN9 D6X0W1 A0A0J9R4W9 A0A0J9R6E4 B3NKS8
PDB
5KUF
E-value=9.22456e-117,
Score=1078
Ontologies
GO
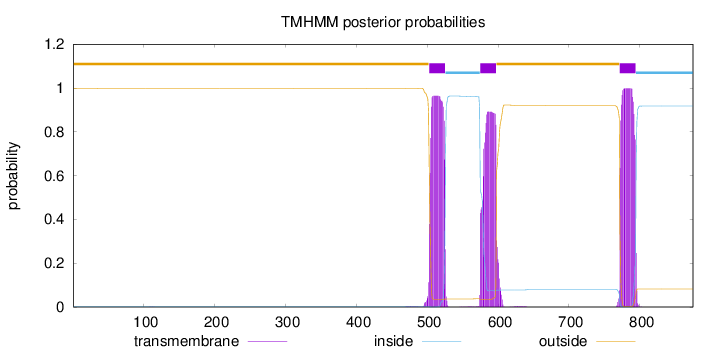
Topology
Subcellular location
Length:
875
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
62.7383
Exp number, first 60 AAs:
0.00115
Total prob of N-in:
0.00294
outside
1 - 502
TMhelix
503 - 525
inside
526 - 574
TMhelix
575 - 597
outside
598 - 771
TMhelix
772 - 794
inside
795 - 875
Population Genetic Test Statistics
Pi
243.025075
Theta
197.361751
Tajima's D
0.807731
CLR
0.528169
CSRT
0.60141992900355
Interpretation
Uncertain
