Gene
KWMTBOMO13760
Pre Gene Modal
BGIBMGA011529
Annotation
PREDICTED:_glutamate_receptor_ionotropic?_kainate_2-like_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.572
Sequence
CDS
ATGTGGGGCACCATGGAATCCGCTCGTCCTTCGGTTTTCGTAAAGAACAACGACGAAGGCGTCGAAAGAGTACTTAAAAGCAGAAGAGGTTATGCCTATTTCATGGAATCAACTGCTATTGAATACCAGTTAGAGCGTCATTGTGAACTAATGCAGATCGGCGACCCTCTTGATTCAAAAGGATACGGAATCGCAATGCCGTTTTTCTCTTCATACCGTGGTGCCGTCGATAACGCTGTACTAAAATTAGCCGAAAGCGGGAAATTGGTGGAGATAAAAAATCGTTGGTGGAAATTGCCTCCCGAAGAAGCTTGTGTTGTTAGTTTTTAA
Protein
MWGTMESARPSVFVKNNDEGVERVLKSRRGYAYFMESTAIEYQLERHCELMQIGDPLDSKGYGIAMPFFSSYRGAVDNAVLKLAESGKLVEIKNRWWKLPPEEACVVSF
Summary
Similarity
Belongs to the glutamate-gated ion channel (TC 1.A.10.1) family.
Uniprot
H9JPS4
A0A2W1BMM8
A0A2H1W6P8
A0A2A4J734
A0A345BF45
A0A194RPP3
+ More
A0A0N1PG13 A0A3S2NWQ7 A0A212EQW1 T1H819 A0A0A9W0D8 A0A0A9XMV4 A0A0A9XKR2 A0A0A9XVL7 A0A0K8T8I2 A0A0L7LUJ9 A0A2R7WSB7 A0A2R7WSA1 A0A1B6H573 A0A1B6JNX3 E0VK11 A0A1B6DA51 A0A3Q0ITI1 A0A158NUC9 A0A1B6DEF0 W5JDY5 A0A2S2PLM7 A0A1B6H180 A0A182TV70 A0A182FK06 A0A182X5A0 A0A182VMN8 A0A182VV99 A0A182I9N5 A0A182RJ06 Q7QE65 A0A182Q917 A0A182M854 A0A182KFG5 A0A182P835 A0A182N102 A0A182YEZ2 A0A182TWK6 A0A182JKW8 A0A2M4BZ21 A0A2M4BZ03 A0A195BSB9 A0A084WPK0 A0A0A1XPV5 A0A140CQC2 A0A182SDM9 A0A2S2R3Y9 A0A2J7QQG2 A0A087ZD04 A0A182YEZ6 A0A182LIW7 W5JDY0 Q7QHV3 A0A182XPJ9 A0A182HFV7 A0A182VIJ3 A0A182RJ09 A0A182VED6 A0A182VVA3 A0A182IZJ3 A0A182U976 T1H541 A0A182N0Z9 A0A2J7QQG7 A0A034V5K6 K7J6E6 A0A1Q3FZA3 B0XGR9 A0A1Q3FZ95 A0A0K8V3R8 A0A182H4S5 A0A1S4F219 Q17HY5 A0A182GWX5 A0A2W1BI66 A0A2P9JYA1 A0A2S2PM36 A0A2H8U1C3 A0A1S5VFU4 A0A1S4F1W5 Q17HY6 A0A2U9NJG4 A0A182H4S6 A0A182G7V3 A0A2J7QQH3 A0A195BLH4 A0A2J7QQK8 A0A2J7QQK1 A0A2J7QQK6 A0A067QWX4 A0A310S5S6 A0A310S9P8 A0A1J1IX76 A0A2R7WBZ6 A0A182P838
A0A0N1PG13 A0A3S2NWQ7 A0A212EQW1 T1H819 A0A0A9W0D8 A0A0A9XMV4 A0A0A9XKR2 A0A0A9XVL7 A0A0K8T8I2 A0A0L7LUJ9 A0A2R7WSB7 A0A2R7WSA1 A0A1B6H573 A0A1B6JNX3 E0VK11 A0A1B6DA51 A0A3Q0ITI1 A0A158NUC9 A0A1B6DEF0 W5JDY5 A0A2S2PLM7 A0A1B6H180 A0A182TV70 A0A182FK06 A0A182X5A0 A0A182VMN8 A0A182VV99 A0A182I9N5 A0A182RJ06 Q7QE65 A0A182Q917 A0A182M854 A0A182KFG5 A0A182P835 A0A182N102 A0A182YEZ2 A0A182TWK6 A0A182JKW8 A0A2M4BZ21 A0A2M4BZ03 A0A195BSB9 A0A084WPK0 A0A0A1XPV5 A0A140CQC2 A0A182SDM9 A0A2S2R3Y9 A0A2J7QQG2 A0A087ZD04 A0A182YEZ6 A0A182LIW7 W5JDY0 Q7QHV3 A0A182XPJ9 A0A182HFV7 A0A182VIJ3 A0A182RJ09 A0A182VED6 A0A182VVA3 A0A182IZJ3 A0A182U976 T1H541 A0A182N0Z9 A0A2J7QQG7 A0A034V5K6 K7J6E6 A0A1Q3FZA3 B0XGR9 A0A1Q3FZ95 A0A0K8V3R8 A0A182H4S5 A0A1S4F219 Q17HY5 A0A182GWX5 A0A2W1BI66 A0A2P9JYA1 A0A2S2PM36 A0A2H8U1C3 A0A1S5VFU4 A0A1S4F1W5 Q17HY6 A0A2U9NJG4 A0A182H4S6 A0A182G7V3 A0A2J7QQH3 A0A195BLH4 A0A2J7QQK8 A0A2J7QQK1 A0A2J7QQK6 A0A067QWX4 A0A310S5S6 A0A310S9P8 A0A1J1IX76 A0A2R7WBZ6 A0A182P838
Pubmed
EMBL
BABH01015232
KZ150075
PZC73970.1
ODYU01006689
SOQ48759.1
NWSH01002576
+ More
PCG67937.1 MG820698 AXF48869.1 KQ459833 KPJ19823.1 KQ459037 KPJ04257.1 RSAL01000042 RVE50833.1 AGBW02013238 OWR43841.1 ACPB03008552 GBHO01042723 JAG00881.1 GBHO01022290 GDHC01007640 JAG21314.1 JAQ10989.1 GBHO01022287 JAG21317.1 GBHO01022289 GBRD01004119 JAG21315.1 JAG61702.1 GBRD01004120 JAG61701.1 JTDY01000067 KOB79074.1 KK855412 PTY22443.1 PTY22444.1 GECU01037857 JAS69849.1 GECU01006758 JAT00949.1 DS235235 EEB13717.1 GEDC01014721 JAS22577.1 ADTU01026376 ADTU01026377 GEDC01026520 GEDC01013278 JAS10778.1 JAS24020.1 ADMH02001821 ETN61009.1 GGMR01017668 MBY30287.1 GECZ01001371 JAS68398.1 APCN01001012 APCN01001013 AAAB01008847 EAA06913.4 AXCN02001376 AXCM01004371 GGFJ01009020 MBW58161.1 GGFJ01009000 MBW58141.1 KQ976417 KYM89915.1 ATLV01025084 KE525369 KFB52144.1 GBXI01001226 JAD13066.1 KR998364 AMH85983.1 GGMS01015526 MBY84729.1 NEVH01012087 PNF30820.1 GGFL01003458 MBW67636.1 ADMH02001779 ETN61105.1 AAAB01008815 EAA05023.4 APCN01007369 CAQQ02162970 CAQQ02162971 PNF30821.1 GAKP01021897 JAC37055.1 AAZX01000424 AAZX01000429 AAZX01008541 AAZX01011194 AAZX01013279 AAZX01013570 AAZX01013893 AAZX01015619 AAZX01025922 GFDL01002185 JAV32860.1 DS233058 EDS27792.1 GFDL01002179 JAV32866.1 GDHF01018808 GDHF01006355 JAI33506.1 JAI45959.1 JXUM01110194 KQ565389 KXJ71033.1 CH477244 EAT46291.1 JXUM01094236 KQ564114 KXJ72756.1 PZC73971.1 KY817091 AVH87307.1 GGMR01017854 MBY30473.1 GFXV01008185 MBW19990.1 KY445581 AQN78516.1 EAT46290.1 MF385150 AWT23321.1 KXJ71034.1 JXUM01008704 KQ560267 KXJ83447.1 PNF30822.1 KQ976453 KYM85527.1 PNF30868.1 PNF30865.1 PNF30864.1 KK853270 KDR09166.1 GGOB01000004 MCH29400.1 GGOB01000002 MCH29398.1 CVRI01000063 CRL04875.1 KK854577 PTY17048.1
PCG67937.1 MG820698 AXF48869.1 KQ459833 KPJ19823.1 KQ459037 KPJ04257.1 RSAL01000042 RVE50833.1 AGBW02013238 OWR43841.1 ACPB03008552 GBHO01042723 JAG00881.1 GBHO01022290 GDHC01007640 JAG21314.1 JAQ10989.1 GBHO01022287 JAG21317.1 GBHO01022289 GBRD01004119 JAG21315.1 JAG61702.1 GBRD01004120 JAG61701.1 JTDY01000067 KOB79074.1 KK855412 PTY22443.1 PTY22444.1 GECU01037857 JAS69849.1 GECU01006758 JAT00949.1 DS235235 EEB13717.1 GEDC01014721 JAS22577.1 ADTU01026376 ADTU01026377 GEDC01026520 GEDC01013278 JAS10778.1 JAS24020.1 ADMH02001821 ETN61009.1 GGMR01017668 MBY30287.1 GECZ01001371 JAS68398.1 APCN01001012 APCN01001013 AAAB01008847 EAA06913.4 AXCN02001376 AXCM01004371 GGFJ01009020 MBW58161.1 GGFJ01009000 MBW58141.1 KQ976417 KYM89915.1 ATLV01025084 KE525369 KFB52144.1 GBXI01001226 JAD13066.1 KR998364 AMH85983.1 GGMS01015526 MBY84729.1 NEVH01012087 PNF30820.1 GGFL01003458 MBW67636.1 ADMH02001779 ETN61105.1 AAAB01008815 EAA05023.4 APCN01007369 CAQQ02162970 CAQQ02162971 PNF30821.1 GAKP01021897 JAC37055.1 AAZX01000424 AAZX01000429 AAZX01008541 AAZX01011194 AAZX01013279 AAZX01013570 AAZX01013893 AAZX01015619 AAZX01025922 GFDL01002185 JAV32860.1 DS233058 EDS27792.1 GFDL01002179 JAV32866.1 GDHF01018808 GDHF01006355 JAI33506.1 JAI45959.1 JXUM01110194 KQ565389 KXJ71033.1 CH477244 EAT46291.1 JXUM01094236 KQ564114 KXJ72756.1 PZC73971.1 KY817091 AVH87307.1 GGMR01017854 MBY30473.1 GFXV01008185 MBW19990.1 KY445581 AQN78516.1 EAT46290.1 MF385150 AWT23321.1 KXJ71034.1 JXUM01008704 KQ560267 KXJ83447.1 PNF30822.1 KQ976453 KYM85527.1 PNF30868.1 PNF30865.1 PNF30864.1 KK853270 KDR09166.1 GGOB01000004 MCH29400.1 GGOB01000002 MCH29398.1 CVRI01000063 CRL04875.1 KK854577 PTY17048.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000283053
UP000007151
+ More
UP000015103 UP000037510 UP000009046 UP000079169 UP000005205 UP000000673 UP000075902 UP000069272 UP000076407 UP000075903 UP000075920 UP000075840 UP000075900 UP000007062 UP000075886 UP000075883 UP000075881 UP000075885 UP000075884 UP000076408 UP000075880 UP000078540 UP000030765 UP000075901 UP000235965 UP000075882 UP000015102 UP000002358 UP000002320 UP000069940 UP000249989 UP000008820 UP000027135 UP000183832
UP000015103 UP000037510 UP000009046 UP000079169 UP000005205 UP000000673 UP000075902 UP000069272 UP000076407 UP000075903 UP000075920 UP000075840 UP000075900 UP000007062 UP000075886 UP000075883 UP000075881 UP000075885 UP000075884 UP000076408 UP000075880 UP000078540 UP000030765 UP000075901 UP000235965 UP000075882 UP000015102 UP000002358 UP000002320 UP000069940 UP000249989 UP000008820 UP000027135 UP000183832
PRIDE
Interpro
SUPFAM
SSF53822
SSF53822
ProteinModelPortal
H9JPS4
A0A2W1BMM8
A0A2H1W6P8
A0A2A4J734
A0A345BF45
A0A194RPP3
+ More
A0A0N1PG13 A0A3S2NWQ7 A0A212EQW1 T1H819 A0A0A9W0D8 A0A0A9XMV4 A0A0A9XKR2 A0A0A9XVL7 A0A0K8T8I2 A0A0L7LUJ9 A0A2R7WSB7 A0A2R7WSA1 A0A1B6H573 A0A1B6JNX3 E0VK11 A0A1B6DA51 A0A3Q0ITI1 A0A158NUC9 A0A1B6DEF0 W5JDY5 A0A2S2PLM7 A0A1B6H180 A0A182TV70 A0A182FK06 A0A182X5A0 A0A182VMN8 A0A182VV99 A0A182I9N5 A0A182RJ06 Q7QE65 A0A182Q917 A0A182M854 A0A182KFG5 A0A182P835 A0A182N102 A0A182YEZ2 A0A182TWK6 A0A182JKW8 A0A2M4BZ21 A0A2M4BZ03 A0A195BSB9 A0A084WPK0 A0A0A1XPV5 A0A140CQC2 A0A182SDM9 A0A2S2R3Y9 A0A2J7QQG2 A0A087ZD04 A0A182YEZ6 A0A182LIW7 W5JDY0 Q7QHV3 A0A182XPJ9 A0A182HFV7 A0A182VIJ3 A0A182RJ09 A0A182VED6 A0A182VVA3 A0A182IZJ3 A0A182U976 T1H541 A0A182N0Z9 A0A2J7QQG7 A0A034V5K6 K7J6E6 A0A1Q3FZA3 B0XGR9 A0A1Q3FZ95 A0A0K8V3R8 A0A182H4S5 A0A1S4F219 Q17HY5 A0A182GWX5 A0A2W1BI66 A0A2P9JYA1 A0A2S2PM36 A0A2H8U1C3 A0A1S5VFU4 A0A1S4F1W5 Q17HY6 A0A2U9NJG4 A0A182H4S6 A0A182G7V3 A0A2J7QQH3 A0A195BLH4 A0A2J7QQK8 A0A2J7QQK1 A0A2J7QQK6 A0A067QWX4 A0A310S5S6 A0A310S9P8 A0A1J1IX76 A0A2R7WBZ6 A0A182P838
A0A0N1PG13 A0A3S2NWQ7 A0A212EQW1 T1H819 A0A0A9W0D8 A0A0A9XMV4 A0A0A9XKR2 A0A0A9XVL7 A0A0K8T8I2 A0A0L7LUJ9 A0A2R7WSB7 A0A2R7WSA1 A0A1B6H573 A0A1B6JNX3 E0VK11 A0A1B6DA51 A0A3Q0ITI1 A0A158NUC9 A0A1B6DEF0 W5JDY5 A0A2S2PLM7 A0A1B6H180 A0A182TV70 A0A182FK06 A0A182X5A0 A0A182VMN8 A0A182VV99 A0A182I9N5 A0A182RJ06 Q7QE65 A0A182Q917 A0A182M854 A0A182KFG5 A0A182P835 A0A182N102 A0A182YEZ2 A0A182TWK6 A0A182JKW8 A0A2M4BZ21 A0A2M4BZ03 A0A195BSB9 A0A084WPK0 A0A0A1XPV5 A0A140CQC2 A0A182SDM9 A0A2S2R3Y9 A0A2J7QQG2 A0A087ZD04 A0A182YEZ6 A0A182LIW7 W5JDY0 Q7QHV3 A0A182XPJ9 A0A182HFV7 A0A182VIJ3 A0A182RJ09 A0A182VED6 A0A182VVA3 A0A182IZJ3 A0A182U976 T1H541 A0A182N0Z9 A0A2J7QQG7 A0A034V5K6 K7J6E6 A0A1Q3FZA3 B0XGR9 A0A1Q3FZ95 A0A0K8V3R8 A0A182H4S5 A0A1S4F219 Q17HY5 A0A182GWX5 A0A2W1BI66 A0A2P9JYA1 A0A2S2PM36 A0A2H8U1C3 A0A1S5VFU4 A0A1S4F1W5 Q17HY6 A0A2U9NJG4 A0A182H4S6 A0A182G7V3 A0A2J7QQH3 A0A195BLH4 A0A2J7QQK8 A0A2J7QQK1 A0A2J7QQK6 A0A067QWX4 A0A310S5S6 A0A310S9P8 A0A1J1IX76 A0A2R7WBZ6 A0A182P838
PDB
5EHS
E-value=8.12505e-29,
Score=309
Ontologies
GO
Topology
Subcellular location
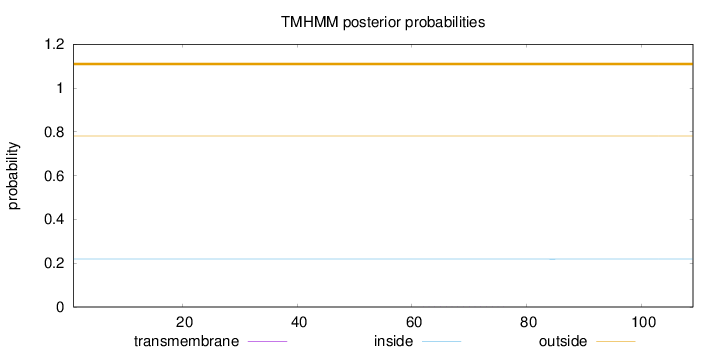
Length:
109
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00319
Exp number, first 60 AAs:
0.00014
Total prob of N-in:
0.21883
outside
1 - 109
Population Genetic Test Statistics
Pi
380.489757
Theta
191.779782
Tajima's D
3.096758
CLR
0
CSRT
0.982250887455627
Interpretation
Uncertain
