Gene
KWMTBOMO13758
Pre Gene Modal
BGIBMGA011528
Annotation
PREDICTED:_glutamate_receptor_ionotropic?_kainate_2_isoform_X2_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 2.316
Sequence
CDS
ATGAGAGGAACAAAAGTCGTTTTCCTCGTACTATTCTTTGGACACCTTTCGGCTCTTCCGGACACGATACGAATAGGTGGACTCTTTCACCCTGAAGATGATAAGCAAGAGGTTGCATTTCGTTACGCAGTTGAAAGAGTTAATGCAGACCGCACCTTACTGCCGCGAGCCAAACTATTAGCCCAAGTCGAAACGATTTCTCCGCAGGACAGTTTTCATGCTTCGAAACGAGTATGTCATCTTCTACGAAGCGGAGTTGCAGCGATATTCGGTCCTCAATCAGCACCCGCTGCCGCTCACGTCCAGTCCATATGCGACACGATGGAACTGCCCCACTTAGAAACCAGATGGGACTATCGCACACGCCGGGAATCTTGCCTCGTAAACCTTTACCCACATCCAGCTGCGCTCAGTAGGGCGTACGTAGATCTTGTTAGAGCTTGGGGTTGGAAATCTTTCACTATAGTATACGAAAATAGCGATGGGTTAGTTCGTCTGCAGGAATTGCTCAAAGCACATGGACCTTCAGAATTGCCTGTATCTGTTCGACAGCTTCCAGATTCTCATGACTATAGACCCCTTCTGAAACAAATAAAAAACTCGGCCGAGTCCCACATTGTTCTCGACTGCGCAACCGAAAGAATAAGGGATGTACTCCAACAAGCACAACAAATCGGAATGATGTCGGATTACCATAGCTACCTTATCACATCTTTGGATCTTCACAGCGTAGATTTGGAAGAGTTTAAATACGGGGGAACAAATATAACTGCTTTAAGACTGCTCGACCCGGAACGAGCAGATGTTCAATGGGTGGTGCGAGATTGGGTTTACGATGAGGCAAGAAAAGGTCGAAAATTGCAATTAGGACACACGACAGCTAAAGAAAATATGACATTTATTAAGACAGAAACAGCCTTGATGTATGACGCAGTACATTTGTTCGCGAAAGCGTTACACGATCTCGATACATCTCAGCAAATCGACGTGAGACCGCTCTCCTGTGAGGCTGAAGATACATGGCCCCATGGGTATAGCCTCATCAATTATATGAAAATCGTCGAAATGAAAGGACTAACCGGAGTTATAAAGTTCGATCACCAAGGATTTAGAAGCGATTTCACTTTAGATATAATCGAATTGACAAGGGACGGTCTCCAAAAAGCTGGAACTTGGAATTCATCTGAAGGTGTCAACTATACGAGATCTTACGGCGAGAACCAAAAACAGATAGTTGAAATTTTGCAAAATAAGACTCTTGTAGTGACAACAATTTTGAGTGCTCCGTATTGTATGAGGAAAGAAGCGAGCGAAAAGTTAACAGGCAACGCACAATTCGAGGGCTACGCAATAGATCTTATTCATGAGATATCTAAAATTCTGGGCTTCAATTATACATTCAAACTGGCTCCTGACGGTCGATACGGGTCGTACAATCGTGAAACCAAAGAATGGGATGGAATGATTAGGGAGCTTCTCGAGCAAAGAGCAGACCTTGCCATAGCAGATCTCACCATTACCTACGACAGAGAGCAAGTGGTCGATTTCACGATGCCATTCATGAACCTCGGCATTTCAGTGCTTTACCGCAAACCAATAAAACAGCCCCCGAACCTCTTCTCATTTCTTTCTCCTTTATCACTGGACGTGTGGATTTATATGGCAACTGCCTATTTAGGGGTTTCTGTGCTGCTGTTTATATTGGCAAGGTTCACTCCATACGAATGGCATCAAACGCATTCACCTGAGGGCGAAAAAATGGAGAATATTTTCTCTCTAGCAAACTGTTTGTGGTTTGCAATCGGATCTCTAATGCAACAAAGCTGTGATTTTTTGCCCAAAGCCGTTTCAACTCGCATGGTGGCCGGGATGTGGTGGTTCTTTACTTTAATCATGATATCATCTTACACTGCCAACTTGGCTGCGTTCTTAACCGTCGAGAGAATGGACTCGCCCATTGAGAGCGCAGAAGATCTCGCAAAACAAACTAAGATCAAATACGGAGCTTTGAAGGGAGGGTCAACTGCAGCATTTTTCAGGGACTCAAACTTTTCAACATACCAGCGAATGTGGTCATTCATGGAATCCGCCAGGCCATCAGTATTCGCAAATACTAACAAGGAGGGAGAAGAAAGGGTGGTGAGGGGAAAGGGCGCTTACGCTTATCTAATGGAGTCTACGACCATAGAATATGTGGTGGAAAGGAACTGTGATCTAACGCAAGTCGGAGGGATGCTCGACTCTAAAGGATATGGAATTGCTATGCCACCAAACTCGCCGTATCGCACAGCCATAAGCGGGGCTGTTTTGAAGCTTCAAGAAGAAGGAAAATTGCACATTCTCAAAACGAAATGGTGGAAGGAAAAACGCGGTGGAGGTTCATGCAGGGATGAAACTTCCAAGTCCTCGTCAACTGCAAACGAGCTAGGTCTAGCCAACGTGGGTGGAGTTTTCGTTGTCCTCATGGGAGGAATGGGAGTAGCATGCGTGATCGCGGTTTGCGAATTCGTATGGAAGTCACGTAAAGTAGCCGTCGATGAACGGAAAGAGGAGGCGTCACTATGCTCGGAGATGGCGTCTGAATTGCGTTCGGCGCTCAAGTGCCCTGGTAGCGCGGGTGGGGGCAGTGGCAGCGGGGCAGCGCGGGATGGCGCGGGTTCCCCATACCTTCACTACGGGTTCAGTACAAAGAGCCAACTCCACTAG
Protein
MRGTKVVFLVLFFGHLSALPDTIRIGGLFHPEDDKQEVAFRYAVERVNADRTLLPRAKLLAQVETISPQDSFHASKRVCHLLRSGVAAIFGPQSAPAAAHVQSICDTMELPHLETRWDYRTRRESCLVNLYPHPAALSRAYVDLVRAWGWKSFTIVYENSDGLVRLQELLKAHGPSELPVSVRQLPDSHDYRPLLKQIKNSAESHIVLDCATERIRDVLQQAQQIGMMSDYHSYLITSLDLHSVDLEEFKYGGTNITALRLLDPERADVQWVVRDWVYDEARKGRKLQLGHTTAKENMTFIKTETALMYDAVHLFAKALHDLDTSQQIDVRPLSCEAEDTWPHGYSLINYMKIVEMKGLTGVIKFDHQGFRSDFTLDIIELTRDGLQKAGTWNSSEGVNYTRSYGENQKQIVEILQNKTLVVTTILSAPYCMRKEASEKLTGNAQFEGYAIDLIHEISKILGFNYTFKLAPDGRYGSYNRETKEWDGMIRELLEQRADLAIADLTITYDREQVVDFTMPFMNLGISVLYRKPIKQPPNLFSFLSPLSLDVWIYMATAYLGVSVLLFILARFTPYEWHQTHSPEGEKMENIFSLANCLWFAIGSLMQQSCDFLPKAVSTRMVAGMWWFFTLIMISSYTANLAAFLTVERMDSPIESAEDLAKQTKIKYGALKGGSTAAFFRDSNFSTYQRMWSFMESARPSVFANTNKEGEERVVRGKGAYAYLMESTTIEYVVERNCDLTQVGGMLDSKGYGIAMPPNSPYRTAISGAVLKLQEEGKLHILKTKWWKEKRGGGSCRDETSKSSSTANELGLANVGGVFVVLMGGMGVACVIAVCEFVWKSRKVAVDERKEEASLCSEMASELRSALKCPGSAGGGSGSGAARDGAGSPYLHYGFSTKSQLH
Summary
Similarity
Belongs to the glutamate-gated ion channel (TC 1.A.10.1) family.
Uniprot
A0A345BF16
A0A2H1WGZ3
A0A0N1I591
A0A0F7QEG0
A0A3S2NLN9
A0A0L7LQ72
+ More
A0A075T818 A0A2A4J849 A0A223HCX9 A0A194RUK6 H9JPS3 A0A0V0J2K9 A0A2J7QQK9 A0A2J7QQL5 A0A2J7QQL0 A0A182FK05 A0A1S4G9Z3 A0A0M4JB15 A0A158NJ03 A0A1W4XFF8 A0A1S4GA38 A0A3L8DZQ4 A0A1Y9GKF3 A0A139WN33 Q7PRP7 A0A1W4XE85 A0A084WPK1 A0A2M4BDE6 F5HJZ1 A0A1W4XF50 A0A1W4X4N7 A0A088ACV5 A0A2A4J773 A0A1Y1KUX6 A0A139WN75 A0A023F5M1 T1H7P3 A0A195DUH1 A0A195FIP6 A0A195D3G4 A0A1S4EBS0 A0A1S4EC64 A0A2C9H8A3 B4LWJ1 B4JUF1 B4K8X7 B4NFR8 Q9VDH5 B4PPU2 B3M354 B4II60 B3P313 A0A1W4VBA5 Q8MS48 A0A3B0KPF1 B4G529 Q299E3 T1PA03 A0A1I8P1D6 W8BH14 A0A1A9V4Z6 A0A0K8UBA7 A0A0A1XQS4 J9JWC6 A0A0M3QXZ2 A0A1A9X5X4 A0A1B0AWM1 A0A2H8TTS0 A0A1B0FK80 B4R1Q6 A0A1A9Z7D1 A0A0L0C0C3 A0A1A9W569 A0A2R7WBG8 A0A026WEA9 A0A034W605 A0A2R7WBZ6 A0A2H8TFU0 E0VYS6 A0A195BBT6 A0A1L2YXN5 E0VK11 A0A2S2PWS9 A0A0P5VYZ8 A0A0P6BPL8 E9FS57 A0A0P5Y8A0 A0A0P6GD71 A0A0P5X0X0 A0A0P4YAD7 A0A182Q917 A0A182N102 A0A182M854 A0A182KFG5 A0A182P835
A0A075T818 A0A2A4J849 A0A223HCX9 A0A194RUK6 H9JPS3 A0A0V0J2K9 A0A2J7QQK9 A0A2J7QQL5 A0A2J7QQL0 A0A182FK05 A0A1S4G9Z3 A0A0M4JB15 A0A158NJ03 A0A1W4XFF8 A0A1S4GA38 A0A3L8DZQ4 A0A1Y9GKF3 A0A139WN33 Q7PRP7 A0A1W4XE85 A0A084WPK1 A0A2M4BDE6 F5HJZ1 A0A1W4XF50 A0A1W4X4N7 A0A088ACV5 A0A2A4J773 A0A1Y1KUX6 A0A139WN75 A0A023F5M1 T1H7P3 A0A195DUH1 A0A195FIP6 A0A195D3G4 A0A1S4EBS0 A0A1S4EC64 A0A2C9H8A3 B4LWJ1 B4JUF1 B4K8X7 B4NFR8 Q9VDH5 B4PPU2 B3M354 B4II60 B3P313 A0A1W4VBA5 Q8MS48 A0A3B0KPF1 B4G529 Q299E3 T1PA03 A0A1I8P1D6 W8BH14 A0A1A9V4Z6 A0A0K8UBA7 A0A0A1XQS4 J9JWC6 A0A0M3QXZ2 A0A1A9X5X4 A0A1B0AWM1 A0A2H8TTS0 A0A1B0FK80 B4R1Q6 A0A1A9Z7D1 A0A0L0C0C3 A0A1A9W569 A0A2R7WBG8 A0A026WEA9 A0A034W605 A0A2R7WBZ6 A0A2H8TFU0 E0VYS6 A0A195BBT6 A0A1L2YXN5 E0VK11 A0A2S2PWS9 A0A0P5VYZ8 A0A0P6BPL8 E9FS57 A0A0P5Y8A0 A0A0P6GD71 A0A0P5X0X0 A0A0P4YAD7 A0A182Q917 A0A182N102 A0A182M854 A0A182KFG5 A0A182P835
Pubmed
29727827
26354079
25803580
26227816
25027790
28150741
+ More
19121390 27006164 12364791 26265180 21347285 30249741 18362917 19820115 24438588 28004739 25474469 17994087 27889096 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 15632085 25315136 24495485 25830018 26108605 24508170 25348373 20566863 21292972
19121390 27006164 12364791 26265180 21347285 30249741 18362917 19820115 24438588 28004739 25474469 17994087 27889096 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 15632085 25315136 24495485 25830018 26108605 24508170 25348373 20566863 21292972
EMBL
MG820669
AXF48840.1
ODYU01008610
SOQ52341.1
KQ459037
KPJ04255.1
+ More
LC017781 BAR64795.1 RSAL01000042 RVE50836.1 JTDY01000347 KOB77595.1 KF768733 AIG51927.1 NWSH01002576 PCG67936.1 KY283580 AST36240.1 KQ459833 KPJ19821.1 BABH01015229 GDKB01000032 JAP38464.1 NEVH01012087 PNF30870.1 PNF30867.1 PNF30869.1 AAAB01008847 KP843209 ALD51346.1 ADTU01017309 ADTU01017310 ADTU01017311 ADTU01017312 ADTU01017313 ADTU01017314 ADTU01017315 ADTU01017316 ADTU01017317 ADTU01017318 QOIP01000002 RLU25960.1 APCN01001007 APCN01001008 APCN01001009 APCN01001010 APCN01001011 APCN01001012 KQ971312 KYB29342.1 EAA06836.6 ATLV01025088 ATLV01025089 ATLV01025090 ATLV01025091 ATLV01025092 KE525369 KFB52145.1 GGFJ01001924 MBW51065.1 EGK96607.1 PCG67935.1 GEZM01076945 JAV63525.1 KYB29343.1 GBBI01002254 JAC16458.1 ACPB03008552 KQ980334 KYN16523.1 KQ981523 KYN40142.1 KQ976885 KYN07450.1 CH940650 EDW67656.1 CH916374 EDV91121.1 CH933806 EDW16574.1 CH964251 EDW83135.1 AE014297 AAF55818.1 CM000160 EDW96192.1 CH902617 EDV42454.2 CH480842 EDW49604.1 CH954181 EDV48327.1 AY119103 AAM50963.1 OUUW01000013 SPP87746.1 CH479179 EDW24695.1 CM000070 EAL27760.2 KA645514 AFP60143.1 GAMC01010282 JAB96273.1 GDHF01028684 JAI23630.1 GBXI01000618 JAD13674.1 ABLF02032802 ABLF02032813 ABLF02032814 ABLF02032817 CP012526 ALC46719.1 JXJN01004815 JXJN01004816 GFXV01005416 MBW17221.1 CCAG010014365 CM000364 EDX12165.1 JRES01001168 KNC24884.1 KK854577 PTY17047.1 KK107250 EZA54440.1 GAKP01007921 JAC51031.1 PTY17048.1 GFXV01001172 MBW12977.1 DS235846 EEB18532.1 KQ976530 KYM81659.1 KX016774 APF30023.1 DS235235 EEB13717.1 GGMS01000159 MBY69362.1 GDIP01167678 GDIP01142972 GDIP01107730 GDIP01059632 LRGB01000848 JAL95984.1 KZS15806.1 GDIP01013465 JAM90250.1 GL732523 EFX90374.1 GDIP01061478 JAM42237.1 GDIQ01035005 JAN59732.1 GDIP01079329 JAM24386.1 GDIP01230363 JAI93038.1 AXCN02001376 AXCM01004371
LC017781 BAR64795.1 RSAL01000042 RVE50836.1 JTDY01000347 KOB77595.1 KF768733 AIG51927.1 NWSH01002576 PCG67936.1 KY283580 AST36240.1 KQ459833 KPJ19821.1 BABH01015229 GDKB01000032 JAP38464.1 NEVH01012087 PNF30870.1 PNF30867.1 PNF30869.1 AAAB01008847 KP843209 ALD51346.1 ADTU01017309 ADTU01017310 ADTU01017311 ADTU01017312 ADTU01017313 ADTU01017314 ADTU01017315 ADTU01017316 ADTU01017317 ADTU01017318 QOIP01000002 RLU25960.1 APCN01001007 APCN01001008 APCN01001009 APCN01001010 APCN01001011 APCN01001012 KQ971312 KYB29342.1 EAA06836.6 ATLV01025088 ATLV01025089 ATLV01025090 ATLV01025091 ATLV01025092 KE525369 KFB52145.1 GGFJ01001924 MBW51065.1 EGK96607.1 PCG67935.1 GEZM01076945 JAV63525.1 KYB29343.1 GBBI01002254 JAC16458.1 ACPB03008552 KQ980334 KYN16523.1 KQ981523 KYN40142.1 KQ976885 KYN07450.1 CH940650 EDW67656.1 CH916374 EDV91121.1 CH933806 EDW16574.1 CH964251 EDW83135.1 AE014297 AAF55818.1 CM000160 EDW96192.1 CH902617 EDV42454.2 CH480842 EDW49604.1 CH954181 EDV48327.1 AY119103 AAM50963.1 OUUW01000013 SPP87746.1 CH479179 EDW24695.1 CM000070 EAL27760.2 KA645514 AFP60143.1 GAMC01010282 JAB96273.1 GDHF01028684 JAI23630.1 GBXI01000618 JAD13674.1 ABLF02032802 ABLF02032813 ABLF02032814 ABLF02032817 CP012526 ALC46719.1 JXJN01004815 JXJN01004816 GFXV01005416 MBW17221.1 CCAG010014365 CM000364 EDX12165.1 JRES01001168 KNC24884.1 KK854577 PTY17047.1 KK107250 EZA54440.1 GAKP01007921 JAC51031.1 PTY17048.1 GFXV01001172 MBW12977.1 DS235846 EEB18532.1 KQ976530 KYM81659.1 KX016774 APF30023.1 DS235235 EEB13717.1 GGMS01000159 MBY69362.1 GDIP01167678 GDIP01142972 GDIP01107730 GDIP01059632 LRGB01000848 JAL95984.1 KZS15806.1 GDIP01013465 JAM90250.1 GL732523 EFX90374.1 GDIP01061478 JAM42237.1 GDIQ01035005 JAN59732.1 GDIP01079329 JAM24386.1 GDIP01230363 JAI93038.1 AXCN02001376 AXCM01004371
Proteomes
UP000053268
UP000283053
UP000037510
UP000218220
UP000053240
UP000005204
+ More
UP000235965 UP000069272 UP000005205 UP000192223 UP000279307 UP000075840 UP000007266 UP000007062 UP000030765 UP000005203 UP000015103 UP000078492 UP000078541 UP000078542 UP000079169 UP000076407 UP000008792 UP000001070 UP000009192 UP000007798 UP000000803 UP000002282 UP000007801 UP000001292 UP000008711 UP000192221 UP000268350 UP000008744 UP000001819 UP000095301 UP000095300 UP000078200 UP000007819 UP000092553 UP000092443 UP000092460 UP000092444 UP000000304 UP000092445 UP000037069 UP000091820 UP000053097 UP000009046 UP000078540 UP000076858 UP000000305 UP000075886 UP000075884 UP000075883 UP000075881 UP000075885
UP000235965 UP000069272 UP000005205 UP000192223 UP000279307 UP000075840 UP000007266 UP000007062 UP000030765 UP000005203 UP000015103 UP000078492 UP000078541 UP000078542 UP000079169 UP000076407 UP000008792 UP000001070 UP000009192 UP000007798 UP000000803 UP000002282 UP000007801 UP000001292 UP000008711 UP000192221 UP000268350 UP000008744 UP000001819 UP000095301 UP000095300 UP000078200 UP000007819 UP000092553 UP000092443 UP000092460 UP000092444 UP000000304 UP000092445 UP000037069 UP000091820 UP000053097 UP000009046 UP000078540 UP000076858 UP000000305 UP000075886 UP000075884 UP000075883 UP000075881 UP000075885
Interpro
SUPFAM
SSF53822
SSF53822
ProteinModelPortal
A0A345BF16
A0A2H1WGZ3
A0A0N1I591
A0A0F7QEG0
A0A3S2NLN9
A0A0L7LQ72
+ More
A0A075T818 A0A2A4J849 A0A223HCX9 A0A194RUK6 H9JPS3 A0A0V0J2K9 A0A2J7QQK9 A0A2J7QQL5 A0A2J7QQL0 A0A182FK05 A0A1S4G9Z3 A0A0M4JB15 A0A158NJ03 A0A1W4XFF8 A0A1S4GA38 A0A3L8DZQ4 A0A1Y9GKF3 A0A139WN33 Q7PRP7 A0A1W4XE85 A0A084WPK1 A0A2M4BDE6 F5HJZ1 A0A1W4XF50 A0A1W4X4N7 A0A088ACV5 A0A2A4J773 A0A1Y1KUX6 A0A139WN75 A0A023F5M1 T1H7P3 A0A195DUH1 A0A195FIP6 A0A195D3G4 A0A1S4EBS0 A0A1S4EC64 A0A2C9H8A3 B4LWJ1 B4JUF1 B4K8X7 B4NFR8 Q9VDH5 B4PPU2 B3M354 B4II60 B3P313 A0A1W4VBA5 Q8MS48 A0A3B0KPF1 B4G529 Q299E3 T1PA03 A0A1I8P1D6 W8BH14 A0A1A9V4Z6 A0A0K8UBA7 A0A0A1XQS4 J9JWC6 A0A0M3QXZ2 A0A1A9X5X4 A0A1B0AWM1 A0A2H8TTS0 A0A1B0FK80 B4R1Q6 A0A1A9Z7D1 A0A0L0C0C3 A0A1A9W569 A0A2R7WBG8 A0A026WEA9 A0A034W605 A0A2R7WBZ6 A0A2H8TFU0 E0VYS6 A0A195BBT6 A0A1L2YXN5 E0VK11 A0A2S2PWS9 A0A0P5VYZ8 A0A0P6BPL8 E9FS57 A0A0P5Y8A0 A0A0P6GD71 A0A0P5X0X0 A0A0P4YAD7 A0A182Q917 A0A182N102 A0A182M854 A0A182KFG5 A0A182P835
A0A075T818 A0A2A4J849 A0A223HCX9 A0A194RUK6 H9JPS3 A0A0V0J2K9 A0A2J7QQK9 A0A2J7QQL5 A0A2J7QQL0 A0A182FK05 A0A1S4G9Z3 A0A0M4JB15 A0A158NJ03 A0A1W4XFF8 A0A1S4GA38 A0A3L8DZQ4 A0A1Y9GKF3 A0A139WN33 Q7PRP7 A0A1W4XE85 A0A084WPK1 A0A2M4BDE6 F5HJZ1 A0A1W4XF50 A0A1W4X4N7 A0A088ACV5 A0A2A4J773 A0A1Y1KUX6 A0A139WN75 A0A023F5M1 T1H7P3 A0A195DUH1 A0A195FIP6 A0A195D3G4 A0A1S4EBS0 A0A1S4EC64 A0A2C9H8A3 B4LWJ1 B4JUF1 B4K8X7 B4NFR8 Q9VDH5 B4PPU2 B3M354 B4II60 B3P313 A0A1W4VBA5 Q8MS48 A0A3B0KPF1 B4G529 Q299E3 T1PA03 A0A1I8P1D6 W8BH14 A0A1A9V4Z6 A0A0K8UBA7 A0A0A1XQS4 J9JWC6 A0A0M3QXZ2 A0A1A9X5X4 A0A1B0AWM1 A0A2H8TTS0 A0A1B0FK80 B4R1Q6 A0A1A9Z7D1 A0A0L0C0C3 A0A1A9W569 A0A2R7WBG8 A0A026WEA9 A0A034W605 A0A2R7WBZ6 A0A2H8TFU0 E0VYS6 A0A195BBT6 A0A1L2YXN5 E0VK11 A0A2S2PWS9 A0A0P5VYZ8 A0A0P6BPL8 E9FS57 A0A0P5Y8A0 A0A0P6GD71 A0A0P5X0X0 A0A0P4YAD7 A0A182Q917 A0A182N102 A0A182M854 A0A182KFG5 A0A182P835
PDB
5KUF
E-value=0,
Score=2069
Ontologies
GO
Topology
Subcellular location
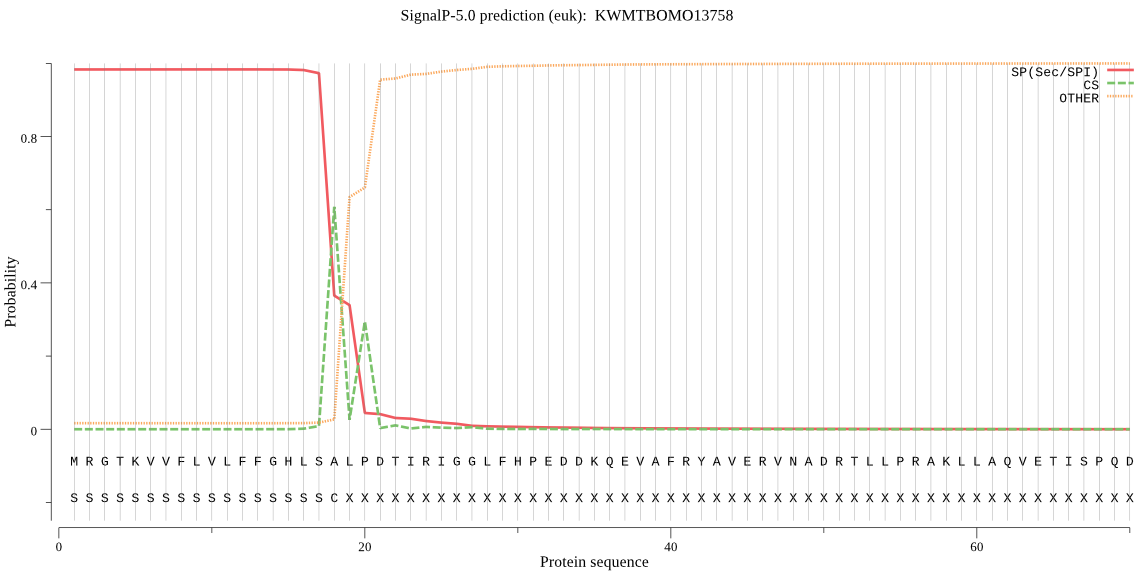
SignalP
Position: 1 - 18,
Likelihood: 0.982933
Length:
901
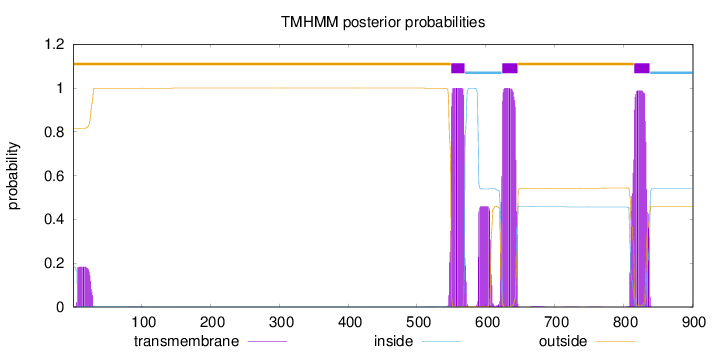
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
78.6203000000002
Exp number, first 60 AAs:
3.77324
Total prob of N-in:
0.18464
outside
1 - 549
TMhelix
550 - 569
inside
570 - 623
TMhelix
624 - 646
outside
647 - 815
TMhelix
816 - 838
inside
839 - 901
Population Genetic Test Statistics
Pi
229.693628
Theta
190.020616
Tajima's D
0.714996
CLR
0.178828
CSRT
0.575821208939553
Interpretation
Uncertain
