Gene
KWMTBOMO13757
Pre Gene Modal
BGIBMGA011486
Annotation
PREDICTED:_glutamate_receptor_ionotropic?_kainate_3-like?_partial_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.711
Sequence
CDS
ATGTGGAATATAGGTTGTAAGGAATCTGATTGTTCGTGGTCTATAATAATTCTAATAAGTCTTCATTTAGTTAATATTACGTGCGGCGACAAAACTATAGGTGCGATTTGCGATGACGGAACTTTCTTATACGAAGCAGCAATTAAAGTTGCAGTCGCTTCAATAACAGAAGAAGAGGAGAGTCCATTGATAGCAAATGTAATACGAACATCTCCCGGCGATTTATTGGAAACGGAAGATGCCGTTTGCTCGTTACTTGAGAAGAATGTTTATGGAATATTTGGGCCGACGAAAAAAGACTCGATCCTCCATGTTCAATCGATCACAGATAATCTAGAAATACCTCAAGTTATAAACCAAGCAATGGAGACACAGAACCGTAATTGGTCAGCAGTAAATCTGTATCCACATCACATCAGCTATTCACAGCTTTTCACAGATCTGATAGAGATGAAGGATTGGAAAGAATTTACTATAATTTATGAGGGCAGCGAACTTTTACCATTTTTGGAAAATATTTTCACTATGCAGGACTTAAACAAAAGACAGAAGATTTTGATGAACGTCGTTCAATTACCCGATGGAGACGATTTCAGGTCTCAGCTGAAAGCAATAAAAAAATCTGGATCAGTTAATTTCGTTTTAGATTGCACTCTCGAAAATTTATCAACGTTTTTAGAACAAGCACAGCAAGTCGGCATAATGTCTGATCAACACAGCTATCTCATAATGAATCCCGATTTCCAAACAATCGATGTCGATCCATTTAAGCATGGAGGTTCGAATATAACAGGTATACGCTTCTTTGATCCTAATCTAGATTCCATTCAGAAGTTCATTTCAGATATAAACGAGAAAGTAGTCGAATTATCCGAAGGTCAGTTAGAAAAAGCTGTTACAGAAAATGGTTTGACTTTTGACTTGGCTTTAATGTATGATGCCGTTACTATTTATGGGTCAGCATTAAACGCACTAGGACTCGAAGAAGGTGCAAATATAACATGTGATCAAGACGACGGCTGGGAGTTTGGTTCTAGCATCATTAATTATATACGAACTATGGAAATTGATGGAATGACGGGAATAATAAAATTCGATGACGAAGGTTTTCGTTCATATTTTGAAGTTGATGTATTAGAAATTATGCCACACGGTTTAGAAAAGGTTGGATCGTGGAATATTGAAGATGGTTTGATACAGGACAGAAATTTTATAGCTCCAGCTGAACCAGAAAGTTCTGAGACGATGAAAGGAAAGCATTTCATTATTTTAACTGCTTTGAGTGCACCCTATGGAATGCTCAAGGAATCATCAAAAAGACTCGAAGGGAACGAACGATACGAAGGATTTGGTATCGAATTAATAGAGGAACTAGCCAAAATGAACGAATTCAATTATACCTTCGAGCTACAAACTGACGGTGTTTATGGATCTTTCGATACAACCACGAAACAGTGGAATGGAATGATGAAGAAAATAATGGACGGTATTGTAGACTTCGGCATAACTGATTTAACGATAACTTCAACACGACAGAAAGCAGTCGACTTTACAAGTCCGTTTATGACGTTAGGAATAACAATATTATACAAAAAACCCACCAAAGAACCACCAGATTTGTTCTCATTTATTTCGCCGCTATCACCAGGAGTTTGGAGCTGGCTAGCGGGAGCTTATATTGGAGTATCAGTTCTGTTATTCGCATTGGGACGAATGGCCCCCGAAGAGTGGCAGAATCCGTACCCGTGTATCGAAGAACCTGAGACCTTGGACAATCAATTCACATTAGCCAATTCCTTTTGGTTTACTCTAGGAAGTGTTCTCACACAAGGATCTGAGATAGCTCCAATAGCCGTTTCAACTCGTATGGCCGGTAGTATGTGGTGGTTCTTCACTTTGATTATGGTGTCGTCCTATACAGCCAACCTCGCCGCTTTCCTAACTATAGAATCAAAGTTTTATGCTATAAAAAGCGTGCAAGATTTAGCCAACAATCCCTACGGTATAACATATGGAGCAAAAAAGAATGGAGCAACATTTAGCTTTTTCATGGAATCCGACAATTTATTGTATCAGAAAATGTACCATTACATGGACGAGCATCCTGAATATCAAACTGCAACGAATGACCAAGGACTTGAAAGAGTAAAATCCGAAAATGAAAATTACGCTTTCCTCATGGAGTCCACATCAATTGAGTATATGGTTGAAAGGAATTGCGACGTTGCTCAAGTCGGCGGCCTATTGGATAACAAAGGATACGGCATTGCCATGAAAAAAAACTCACCTTACCGACAACCGATGAGTGAATCTATATTACAGCTACAGGAAGATGGTACTTTGACTAAATTAAAAGATAAGTGGTGGAAAGAAAAAAGAGGCGGTGGTGCTTGTGAGGATGACGATACCGCTGGCGGTGAAGCCCAACCGTTGGTGTTAGCAAACGTCGGTGGTGTTTTCATAGTTCTAGTGGTCGGCTCAGCAATGGCGGCTGTATGTGCATTTTTCGAGATGTTATTCGATGTTTGGATGATTTGTCGGAGACATAAATTGCCGTTCCTAGATGAATTGAAAGCGGAGTTGAGATTTATTCTGAGTTTTAGTGGAGACACGAAGCCGGTACGTCACCGTGAATTGTCAGGTAGTGGATCACGAAGCACCAAAAACGACGAGGAAGATAAGAATTGTGAATTGGAATCTTTGGATGATGGAAATAAATTGGATCCGGCACCAACGCCGAGATCTGAAAGGTCCACGCATTCACACCACACAATACATAGTAGAAGAACAAGCAATGCAGTTCAAATGGCTAAAATGAGAAAATATAGTATTAGAAGTGGCATATAA
Protein
MWNIGCKESDCSWSIIILISLHLVNITCGDKTIGAICDDGTFLYEAAIKVAVASITEEEESPLIANVIRTSPGDLLETEDAVCSLLEKNVYGIFGPTKKDSILHVQSITDNLEIPQVINQAMETQNRNWSAVNLYPHHISYSQLFTDLIEMKDWKEFTIIYEGSELLPFLENIFTMQDLNKRQKILMNVVQLPDGDDFRSQLKAIKKSGSVNFVLDCTLENLSTFLEQAQQVGIMSDQHSYLIMNPDFQTIDVDPFKHGGSNITGIRFFDPNLDSIQKFISDINEKVVELSEGQLEKAVTENGLTFDLALMYDAVTIYGSALNALGLEEGANITCDQDDGWEFGSSIINYIRTMEIDGMTGIIKFDDEGFRSYFEVDVLEIMPHGLEKVGSWNIEDGLIQDRNFIAPAEPESSETMKGKHFIILTALSAPYGMLKESSKRLEGNERYEGFGIELIEELAKMNEFNYTFELQTDGVYGSFDTTTKQWNGMMKKIMDGIVDFGITDLTITSTRQKAVDFTSPFMTLGITILYKKPTKEPPDLFSFISPLSPGVWSWLAGAYIGVSVLLFALGRMAPEEWQNPYPCIEEPETLDNQFTLANSFWFTLGSVLTQGSEIAPIAVSTRMAGSMWWFFTLIMVSSYTANLAAFLTIESKFYAIKSVQDLANNPYGITYGAKKNGATFSFFMESDNLLYQKMYHYMDEHPEYQTATNDQGLERVKSENENYAFLMESTSIEYMVERNCDVAQVGGLLDNKGYGIAMKKNSPYRQPMSESILQLQEDGTLTKLKDKWWKEKRGGGACEDDDTAGGEAQPLVLANVGGVFIVLVVGSAMAAVCAFFEMLFDVWMICRRHKLPFLDELKAELRFILSFSGDTKPVRHRELSGSGSRSTKNDEEDKNCELESLDDGNKLDPAPTPRSERSTHSHHTIHSRRTSNAVQMAKMRKYSIRSGI
Summary
Similarity
Belongs to the glutamate-gated ion channel (TC 1.A.10.1) family.
Uniprot
H9JPN1
A0A075T675
A0A2A4J8S4
A0A2H1VML2
A0A2W1BQI7
A0A0B5A1Y0
+ More
A0A194RR85 A0A0N0PA90 A0A212ER31 A0A1S4F217 Q17HZ0 Q7QCT5 A0A1Q3G542 A0A1Q3G4Z8 A0A336M7Y1 A0A336M1P8 A0A084WPK2 A0A182VF89 A0A182IWS9 A0A182P839 A0A1S4G9X1 A0A182X5A4 A0A182QKT0 A0A182V8U1 Q5TUR9 A0A1Q3FV55 F5HKL2 A0A182RIF0 A0A1Q3FVD5 A0A182I9N1 A0A182N793 A0A182K4K3 A0A182TY63 A0A182RJ10 A0A182MGS7 A0A1B6DVP7 A0A182VVA4 A0A182YEZ7 A0A182TZ79 A0A336MCA3 E0VYS5 A0A336KBS6 A0A182W0P2 A0A2J7QQF6 A0A2J7QQG5 A0A182FK05 A0A182H5J6 A0A067QJY2 A0A0K8VI05 A0A182PQM6 A0A0A1XKL4 A0A140CQD2 A0A1B6CLB4 A0A140CQC1 A0A034VCG6 T1H862 A0A182LGK9 J9JWC6 A0A182HTL3 A0A0M4JB15 A0A1B6J4X8 W8BXV3 A0A2J7QQL5 A0A0M3SBL8 A0A1J1J1Y8 A0A2J7QQL0 W8BL80 A0A1J1IXR3 A0A0F7QEG0 A0A0G2UKD8 A0A3L8DZQ4 T1H7P3 A0A2J7QQF4 A0A075T818 A0A3S2NLN9 A0A2A4J849 A0A182XHS4 A0A1A9Z7D4 A0A023F5M1 A0A1W4X4N7 A0A2J7QQK9 A0A0K8VYA0 A0A0R1E1K4 A0A0K8V9K4 A0A1W4XE85 A0A2S2Q418 Q0KI38 B3M356 A0A0V0J2K9 A0A0B4KHJ0 A0A154NX52 A0A1S4GA38 B4PQ90 F5HJZ1 A0A2H8TH02 A0A1W4VBC7 T1PEB1 A0A1W4XFF8 A0A3B0K4X3
A0A194RR85 A0A0N0PA90 A0A212ER31 A0A1S4F217 Q17HZ0 Q7QCT5 A0A1Q3G542 A0A1Q3G4Z8 A0A336M7Y1 A0A336M1P8 A0A084WPK2 A0A182VF89 A0A182IWS9 A0A182P839 A0A1S4G9X1 A0A182X5A4 A0A182QKT0 A0A182V8U1 Q5TUR9 A0A1Q3FV55 F5HKL2 A0A182RIF0 A0A1Q3FVD5 A0A182I9N1 A0A182N793 A0A182K4K3 A0A182TY63 A0A182RJ10 A0A182MGS7 A0A1B6DVP7 A0A182VVA4 A0A182YEZ7 A0A182TZ79 A0A336MCA3 E0VYS5 A0A336KBS6 A0A182W0P2 A0A2J7QQF6 A0A2J7QQG5 A0A182FK05 A0A182H5J6 A0A067QJY2 A0A0K8VI05 A0A182PQM6 A0A0A1XKL4 A0A140CQD2 A0A1B6CLB4 A0A140CQC1 A0A034VCG6 T1H862 A0A182LGK9 J9JWC6 A0A182HTL3 A0A0M4JB15 A0A1B6J4X8 W8BXV3 A0A2J7QQL5 A0A0M3SBL8 A0A1J1J1Y8 A0A2J7QQL0 W8BL80 A0A1J1IXR3 A0A0F7QEG0 A0A0G2UKD8 A0A3L8DZQ4 T1H7P3 A0A2J7QQF4 A0A075T818 A0A3S2NLN9 A0A2A4J849 A0A182XHS4 A0A1A9Z7D4 A0A023F5M1 A0A1W4X4N7 A0A2J7QQK9 A0A0K8VYA0 A0A0R1E1K4 A0A0K8V9K4 A0A1W4XE85 A0A2S2Q418 Q0KI38 B3M356 A0A0V0J2K9 A0A0B4KHJ0 A0A154NX52 A0A1S4GA38 B4PQ90 F5HJZ1 A0A2H8TH02 A0A1W4VBC7 T1PEB1 A0A1W4XFF8 A0A3B0K4X3
Pubmed
EMBL
BABH01015227
KF768731
AIG51925.1
NWSH01002576
PCG67934.1
ODYU01003373
+ More
SOQ42048.1 KZ150075 PZC73973.1 KJ542729 AJD81613.1 KQ459833 KPJ19820.1 KQ459037 KPJ04254.1 AGBW02013122 OWR43953.1 CH477244 EAT46286.1 AAAB01008859 EAA07814.5 GFDL01000159 JAV34886.1 GFDL01000165 JAV34880.1 UFQS01000674 UFQT01000674 SSX06068.1 SSX26424.1 UFQS01000248 UFQT01000248 SSX02000.1 SSX22377.1 ATLV01025092 KE525369 KFB52146.1 AAAB01008847 AXCN02001179 EAL41148.4 GFDL01003610 JAV31435.1 EGK96764.1 GFDL01003609 JAV31436.1 APCN01001006 APCN01001007 AXCM01007252 GEDC01007565 JAS29733.1 SSX06067.1 SSX26423.1 DS235846 EEB18531.1 SSX01999.1 SSX22376.1 NEVH01012087 PNF30819.1 PNF30817.1 JXUM01111893 KQ565546 KXJ70888.1 KK853270 KDR09169.1 GDHF01013780 JAI38534.1 GBXI01002423 JAD11869.1 KR998374 AMH85993.1 GEDC01023044 JAS14254.1 KR998363 AMH85982.1 GAKP01019150 JAC39802.1 ACPB03008552 ABLF02032802 ABLF02032813 ABLF02032814 ABLF02032817 APCN01000567 KP843209 ALD51346.1 GECU01013484 JAS94222.1 GAMC01012486 GAMC01012485 JAB94069.1 PNF30867.1 KP843215 ALD51352.1 CVRI01000063 CRL04873.1 PNF30869.1 GAMC01012484 GAMC01012482 JAB94071.1 CRL04872.1 LC017781 BAR64795.1 KP743677 AKI28993.1 QOIP01000002 RLU25960.1 PNF30815.1 KF768733 AIG51927.1 RSAL01000042 RVE50836.1 PCG67936.1 GBBI01002254 JAC16458.1 PNF30870.1 GDHF01008505 JAI43809.1 CM000160 KRK02975.1 GDHF01016670 JAI35644.1 GGMS01003263 MBY72466.1 AE014297 ABI31186.2 CH902617 EDV42456.2 GDKB01000032 JAP38464.1 AGB96168.1 KQ434777 KZC04142.1 EDW96199.1 EGK96607.1 GFXV01001516 MBW13321.1 KA647076 AFP61705.1 OUUW01000013 SPP87752.1
SOQ42048.1 KZ150075 PZC73973.1 KJ542729 AJD81613.1 KQ459833 KPJ19820.1 KQ459037 KPJ04254.1 AGBW02013122 OWR43953.1 CH477244 EAT46286.1 AAAB01008859 EAA07814.5 GFDL01000159 JAV34886.1 GFDL01000165 JAV34880.1 UFQS01000674 UFQT01000674 SSX06068.1 SSX26424.1 UFQS01000248 UFQT01000248 SSX02000.1 SSX22377.1 ATLV01025092 KE525369 KFB52146.1 AAAB01008847 AXCN02001179 EAL41148.4 GFDL01003610 JAV31435.1 EGK96764.1 GFDL01003609 JAV31436.1 APCN01001006 APCN01001007 AXCM01007252 GEDC01007565 JAS29733.1 SSX06067.1 SSX26423.1 DS235846 EEB18531.1 SSX01999.1 SSX22376.1 NEVH01012087 PNF30819.1 PNF30817.1 JXUM01111893 KQ565546 KXJ70888.1 KK853270 KDR09169.1 GDHF01013780 JAI38534.1 GBXI01002423 JAD11869.1 KR998374 AMH85993.1 GEDC01023044 JAS14254.1 KR998363 AMH85982.1 GAKP01019150 JAC39802.1 ACPB03008552 ABLF02032802 ABLF02032813 ABLF02032814 ABLF02032817 APCN01000567 KP843209 ALD51346.1 GECU01013484 JAS94222.1 GAMC01012486 GAMC01012485 JAB94069.1 PNF30867.1 KP843215 ALD51352.1 CVRI01000063 CRL04873.1 PNF30869.1 GAMC01012484 GAMC01012482 JAB94071.1 CRL04872.1 LC017781 BAR64795.1 KP743677 AKI28993.1 QOIP01000002 RLU25960.1 PNF30815.1 KF768733 AIG51927.1 RSAL01000042 RVE50836.1 PCG67936.1 GBBI01002254 JAC16458.1 PNF30870.1 GDHF01008505 JAI43809.1 CM000160 KRK02975.1 GDHF01016670 JAI35644.1 GGMS01003263 MBY72466.1 AE014297 ABI31186.2 CH902617 EDV42456.2 GDKB01000032 JAP38464.1 AGB96168.1 KQ434777 KZC04142.1 EDW96199.1 EGK96607.1 GFXV01001516 MBW13321.1 KA647076 AFP61705.1 OUUW01000013 SPP87752.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000008820
+ More
UP000007062 UP000030765 UP000075903 UP000075880 UP000075885 UP000076407 UP000075886 UP000075900 UP000075840 UP000075884 UP000075881 UP000075902 UP000075883 UP000075920 UP000076408 UP000009046 UP000235965 UP000069272 UP000069940 UP000249989 UP000027135 UP000015103 UP000075882 UP000007819 UP000183832 UP000279307 UP000283053 UP000092445 UP000192223 UP000002282 UP000000803 UP000007801 UP000076502 UP000192221 UP000268350
UP000007062 UP000030765 UP000075903 UP000075880 UP000075885 UP000076407 UP000075886 UP000075900 UP000075840 UP000075884 UP000075881 UP000075902 UP000075883 UP000075920 UP000076408 UP000009046 UP000235965 UP000069272 UP000069940 UP000249989 UP000027135 UP000015103 UP000075882 UP000007819 UP000183832 UP000279307 UP000283053 UP000092445 UP000192223 UP000002282 UP000000803 UP000007801 UP000076502 UP000192221 UP000268350
Interpro
ProteinModelPortal
H9JPN1
A0A075T675
A0A2A4J8S4
A0A2H1VML2
A0A2W1BQI7
A0A0B5A1Y0
+ More
A0A194RR85 A0A0N0PA90 A0A212ER31 A0A1S4F217 Q17HZ0 Q7QCT5 A0A1Q3G542 A0A1Q3G4Z8 A0A336M7Y1 A0A336M1P8 A0A084WPK2 A0A182VF89 A0A182IWS9 A0A182P839 A0A1S4G9X1 A0A182X5A4 A0A182QKT0 A0A182V8U1 Q5TUR9 A0A1Q3FV55 F5HKL2 A0A182RIF0 A0A1Q3FVD5 A0A182I9N1 A0A182N793 A0A182K4K3 A0A182TY63 A0A182RJ10 A0A182MGS7 A0A1B6DVP7 A0A182VVA4 A0A182YEZ7 A0A182TZ79 A0A336MCA3 E0VYS5 A0A336KBS6 A0A182W0P2 A0A2J7QQF6 A0A2J7QQG5 A0A182FK05 A0A182H5J6 A0A067QJY2 A0A0K8VI05 A0A182PQM6 A0A0A1XKL4 A0A140CQD2 A0A1B6CLB4 A0A140CQC1 A0A034VCG6 T1H862 A0A182LGK9 J9JWC6 A0A182HTL3 A0A0M4JB15 A0A1B6J4X8 W8BXV3 A0A2J7QQL5 A0A0M3SBL8 A0A1J1J1Y8 A0A2J7QQL0 W8BL80 A0A1J1IXR3 A0A0F7QEG0 A0A0G2UKD8 A0A3L8DZQ4 T1H7P3 A0A2J7QQF4 A0A075T818 A0A3S2NLN9 A0A2A4J849 A0A182XHS4 A0A1A9Z7D4 A0A023F5M1 A0A1W4X4N7 A0A2J7QQK9 A0A0K8VYA0 A0A0R1E1K4 A0A0K8V9K4 A0A1W4XE85 A0A2S2Q418 Q0KI38 B3M356 A0A0V0J2K9 A0A0B4KHJ0 A0A154NX52 A0A1S4GA38 B4PQ90 F5HJZ1 A0A2H8TH02 A0A1W4VBC7 T1PEB1 A0A1W4XFF8 A0A3B0K4X3
A0A194RR85 A0A0N0PA90 A0A212ER31 A0A1S4F217 Q17HZ0 Q7QCT5 A0A1Q3G542 A0A1Q3G4Z8 A0A336M7Y1 A0A336M1P8 A0A084WPK2 A0A182VF89 A0A182IWS9 A0A182P839 A0A1S4G9X1 A0A182X5A4 A0A182QKT0 A0A182V8U1 Q5TUR9 A0A1Q3FV55 F5HKL2 A0A182RIF0 A0A1Q3FVD5 A0A182I9N1 A0A182N793 A0A182K4K3 A0A182TY63 A0A182RJ10 A0A182MGS7 A0A1B6DVP7 A0A182VVA4 A0A182YEZ7 A0A182TZ79 A0A336MCA3 E0VYS5 A0A336KBS6 A0A182W0P2 A0A2J7QQF6 A0A2J7QQG5 A0A182FK05 A0A182H5J6 A0A067QJY2 A0A0K8VI05 A0A182PQM6 A0A0A1XKL4 A0A140CQD2 A0A1B6CLB4 A0A140CQC1 A0A034VCG6 T1H862 A0A182LGK9 J9JWC6 A0A182HTL3 A0A0M4JB15 A0A1B6J4X8 W8BXV3 A0A2J7QQL5 A0A0M3SBL8 A0A1J1J1Y8 A0A2J7QQL0 W8BL80 A0A1J1IXR3 A0A0F7QEG0 A0A0G2UKD8 A0A3L8DZQ4 T1H7P3 A0A2J7QQF4 A0A075T818 A0A3S2NLN9 A0A2A4J849 A0A182XHS4 A0A1A9Z7D4 A0A023F5M1 A0A1W4X4N7 A0A2J7QQK9 A0A0K8VYA0 A0A0R1E1K4 A0A0K8V9K4 A0A1W4XE85 A0A2S2Q418 Q0KI38 B3M356 A0A0V0J2K9 A0A0B4KHJ0 A0A154NX52 A0A1S4GA38 B4PQ90 F5HJZ1 A0A2H8TH02 A0A1W4VBC7 T1PEB1 A0A1W4XFF8 A0A3B0K4X3
PDB
5KUF
E-value=1.59355e-158,
Score=1439
Ontologies
GO
Topology
Subcellular location
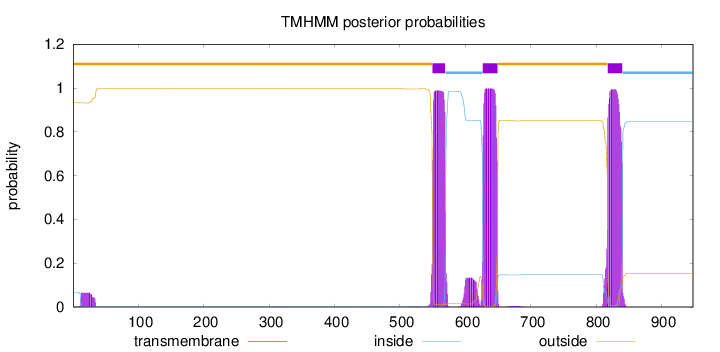
Length:
948
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
70.1369900000001
Exp number, first 60 AAs:
1.36427
Total prob of N-in:
0.06734
outside
1 - 549
TMhelix
550 - 569
inside
570 - 626
TMhelix
627 - 649
outside
650 - 817
TMhelix
818 - 840
inside
841 - 948
Population Genetic Test Statistics
Pi
219.921184
Theta
177.450055
Tajima's D
0.741901
CLR
0
CSRT
0.584320783960802
Interpretation
Uncertain
