Pre Gene Modal
BGIBMGA011489
Annotation
PREDICTED:_monocarboxylate_transporter_10-like_isoform_X2_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 4.321
Sequence
CDS
ATGGAAGAGTCATGGTCGTCGGGCGGCGCGCCGCTGCTGCGCACCAGGTCTCTACCGCTGGTGCTGGAGGCGGCGAGTGGAGCACCCGCGCGCCAGGCGAAGCTCGATCCCCGTCAGCTACTCACGCTCACGCGACATTATTATCCCGAGGGCGGCTGGGGATGGGGGGTAACCGCCGCGGCCACTCTAGCGCAACTCTTGGCGCATGGCCTCCACCAGGCGTCGGCTATTCTAGCGGTAGAAGCTATAAGGCGGTTCGGCCCAGAAGTCCGAATGCAAGCAGGATGTCTAGGTGCTTTGTCAGCGGGGGTGGCACTGGCCTTGTCTCCAGTAACAGTGGCCCTCTGCGTTCGAAAGTCTACACGCGTTACGGCGGTGGTGGGTGGTTTAGTAGCAGCTCTGGGATGTCTCTTCACATCATTTGCCACACAATTTCATCAGCTGTTTTTCAGTTATGGAACGGTGGTAGGTGTGGGCGTGGGTCTCACGAGAGATTGTTCGACTTTAATGGTAGCTCAATACTTCAAAAGAAGACGGGAGCTGGTAGAAATATTTATAGTCAGCGGCAGCGGTCTGGGCATAGCTGTCATGTCAACGTTCATCAAAGGTGTCATCAGAGCGATCGGGTGGCGACTTGGCTTGCAGGCAGTCACCGGAGTAGTATTCGTTACCTTTATTCTGGGAACATTCTACCGTTCGGCTTCTCTGTATCATCCCCAGCGACGAGCAATTTTACATTTGAAGAACCAGAATAAGATCAAGAGGAAAATGAAAGACAGGCAGAAAGCAGACGATAGACAACCGTTCTTCGATTTCTCGACGCTTAAATCGAAAACAGTCAGAATCTTGCTAATGTCAACTGGAATTTCCGCGTTCGGAATAAACACGCCTATATTTTATTTAGCCTATCACGCTGAGGAAGAAGGTTTAGGTGACACAGCTGAACTGCTCCAGGCTTATTTGGGACTCGCCTGGGCAGTGGGCTGTGCAGCGTTCGGCCTGCTAGTCCGACAAAACTCGGCCGAATGCAGAATCGCTAGGCAGTATCTAACTCAAGCAGCTGTTTTCGTTTGTGGACTAGCTACTATGGCACTCACGGCTGTCGAAGGATCCTATAGGGGCTATGTTATGTTCGCTTGGGTATACGGGATTTTCTGCGGTGGCTACCATTACTCTTTGAAGATGTATACCTACGAAAGAGTCAGGTCACGAAATTTTGCACGCACTTGGGGCTTCGTGCAATGCTCTCAAGCGGTGCCCATTGCTATTGGAGTTCCTTTATCAGGTTACATTAACGTTGGCTGCGGCGGCAAGGCGGGCTATTACTTCAGCTCAACGTGTTCTCTTATCGGCTCTCTGTCTCTCTTCTGTATAGACCTGCATCGTCGCAGCGTCGCTCATAAACACACCAAAGAAAATGGCGGTACGGCTACATGCGAATCGGCGTGTGGACCTCGTCGCAGCCGCGAGCGAGAACCGCGCACCGCCGTGGGAGCTGCTACCGCTTTAGTTCTAGGAGCAGAATTGATAGCCCCTGATAGGGCCGGTCGAGATCTACTAGATAGCATAGGTCCAGGAAGCCTCGGGTCACCGCCACCTAATGTTCCTCCTGAACTAACCTGCATCAGCGAAGAAGGTGGCCTTGATCTCGACTTGGATTTGGAGATCCCAGAGCACCTTCTCGAAGACCTGGACTGCGGTGGCGATTGTATCACAAGTTGTAATAAGGTGGAAAACTATTTAATGTTGAGCGAATACGAAAACAATCTGATAGCTGAACTGCCGAATTTGAACGAACGTCGCGGGAGGCGGTGGTCCGTCGTCGTCGGGAATTCCCCTCCGCAGCCATCCTCTATACACGAAGAACAATCGCCCGAGAACAACTCCAAGAACGCGAAATGGAAGAAAAAGTGCCATAACAACACAAACAACAGACTGATTACGGTGATCAACGAGGCGTCACTTTAA
Protein
MEESWSSGGAPLLRTRSLPLVLEAASGAPARQAKLDPRQLLTLTRHYYPEGGWGWGVTAAATLAQLLAHGLHQASAILAVEAIRRFGPEVRMQAGCLGALSAGVALALSPVTVALCVRKSTRVTAVVGGLVAALGCLFTSFATQFHQLFFSYGTVVGVGVGLTRDCSTLMVAQYFKRRRELVEIFIVSGSGLGIAVMSTFIKGVIRAIGWRLGLQAVTGVVFVTFILGTFYRSASLYHPQRRAILHLKNQNKIKRKMKDRQKADDRQPFFDFSTLKSKTVRILLMSTGISAFGINTPIFYLAYHAEEEGLGDTAELLQAYLGLAWAVGCAAFGLLVRQNSAECRIARQYLTQAAVFVCGLATMALTAVEGSYRGYVMFAWVYGIFCGGYHYSLKMYTYERVRSRNFARTWGFVQCSQAVPIAIGVPLSGYINVGCGGKAGYYFSSTCSLIGSLSLFCIDLHRRSVAHKHTKENGGTATCESACGPRRSREREPRTAVGAATALVLGAELIAPDRAGRDLLDSIGPGSLGSPPPNVPPELTCISEEGGLDLDLDLEIPEHLLEDLDCGGDCITSCNKVENYLMLSEYENNLIAELPNLNERRGRRWSVVVGNSPPQPSSIHEEQSPENNSKNAKWKKKCHNNTNNRLITVINEASL
Summary
Uniprot
A0A2A4IW41
A0A212FAI9
A0A194PL08
H9JPN4
A0A2W1BL11
A0A194RQQ6
+ More
A0A088ACW2 A0A154NYQ3 A0A195FIP1 A0A0L7QPC4 F4WWQ8 A0A3L8DYK7 A0A158NJ06 A0A195BL72 E2BH66 A0A151JN04 A0A1W4X8F4 A0A139WN08 A0A1B6KVF5 A0A0L7LU93 A0A1B6CYN4 A0A1W4X8H0 A0A1W4XIT9 A0A2J7QPW8 Q16M19 A0A1S4EGT1 A0A182GH87 A0A084WPL5 B0WHV8 F5HJZ0 A0A310SIM6 A0A182FJZ9 A0A182M0I4 A0A023F483 A0A182YF02 A0A182Q1B4 A0A182VVB2 A0A182RJ17 A0A182VKW5 A7USK6 A0A182J697 A0A182PM63 A0A182N0Z2 A0A182KFP7 A0A182I9M4 W5JQ81 A0A182SPN8 A0A182X5B1 A0A182TKB4 A0A336KAV3 F5HJY9 A0A336LWF0 T1I7F8 A0A1Y1L5L1 A0A336MCK4 A0A1Y1L844 A0A1B0EVG7 N6U5G7 A0A0J7P5H8 A0A0A9W6V5 A0A1J1IZ92 E0VYS1 A0A226DYV1 T1J6X6 E9GUQ3 A0A226DWW9
A0A088ACW2 A0A154NYQ3 A0A195FIP1 A0A0L7QPC4 F4WWQ8 A0A3L8DYK7 A0A158NJ06 A0A195BL72 E2BH66 A0A151JN04 A0A1W4X8F4 A0A139WN08 A0A1B6KVF5 A0A0L7LU93 A0A1B6CYN4 A0A1W4X8H0 A0A1W4XIT9 A0A2J7QPW8 Q16M19 A0A1S4EGT1 A0A182GH87 A0A084WPL5 B0WHV8 F5HJZ0 A0A310SIM6 A0A182FJZ9 A0A182M0I4 A0A023F483 A0A182YF02 A0A182Q1B4 A0A182VVB2 A0A182RJ17 A0A182VKW5 A7USK6 A0A182J697 A0A182PM63 A0A182N0Z2 A0A182KFP7 A0A182I9M4 W5JQ81 A0A182SPN8 A0A182X5B1 A0A182TKB4 A0A336KAV3 F5HJY9 A0A336LWF0 T1I7F8 A0A1Y1L5L1 A0A336MCK4 A0A1Y1L844 A0A1B0EVG7 N6U5G7 A0A0J7P5H8 A0A0A9W6V5 A0A1J1IZ92 E0VYS1 A0A226DYV1 T1J6X6 E9GUQ3 A0A226DWW9
Pubmed
EMBL
NWSH01005465
PCG64187.1
AGBW02009469
OWR50752.1
KQ459601
KPI94017.1
+ More
BABH01015213 KZ150075 PZC73977.1 KQ459833 KPJ19817.1 KQ434777 KZC04148.1 KQ981523 KYN40137.1 KQ414830 KOC60404.1 GL888412 EGI61380.1 QOIP01000002 RLU25517.1 ADTU01017340 ADTU01017341 ADTU01017342 KQ976453 KYM85519.1 GL448268 EFN84987.1 KQ978889 KYN27665.1 KQ971312 KYB29195.1 GEBQ01024564 JAT15413.1 JTDY01000070 KOB79048.1 GEDC01018787 JAS18511.1 NEVH01012087 PNF30630.1 CH477884 EAT35364.1 JXUM01001281 JXUM01001282 JXUM01001283 JXUM01001284 JXUM01001285 KQ560119 KXJ84383.1 ATLV01025100 ATLV01025101 KE525369 KFB52159.1 DS231940 EDS28018.1 AAAB01008847 EGK96605.1 KQ759841 OAD62628.1 AXCM01007783 GBBI01002406 JAC16306.1 AXCN02001373 EDO64304.2 APCN01001005 ADMH02000784 ETN65060.1 UFQS01000248 UFQT01000248 SSX02006.1 SSX22383.1 EGK96606.1 SSX02005.1 SSX22382.1 ACPB03006290 GEZM01063783 JAV68959.1 UFQT01000851 SSX27650.1 GEZM01063784 JAV68958.1 AJWK01020210 APGK01048498 KB741112 KB632184 ENN73817.1 ERL89632.1 LBMM01000050 KMR05159.1 GBHO01039432 GBHO01039429 JAG04172.1 JAG04175.1 CVRI01000063 CRL04860.1 DS235846 EEB18527.1 LNIX01000009 OXA49974.1 JH431897 GL732566 EFX76773.1 OXA49975.1
BABH01015213 KZ150075 PZC73977.1 KQ459833 KPJ19817.1 KQ434777 KZC04148.1 KQ981523 KYN40137.1 KQ414830 KOC60404.1 GL888412 EGI61380.1 QOIP01000002 RLU25517.1 ADTU01017340 ADTU01017341 ADTU01017342 KQ976453 KYM85519.1 GL448268 EFN84987.1 KQ978889 KYN27665.1 KQ971312 KYB29195.1 GEBQ01024564 JAT15413.1 JTDY01000070 KOB79048.1 GEDC01018787 JAS18511.1 NEVH01012087 PNF30630.1 CH477884 EAT35364.1 JXUM01001281 JXUM01001282 JXUM01001283 JXUM01001284 JXUM01001285 KQ560119 KXJ84383.1 ATLV01025100 ATLV01025101 KE525369 KFB52159.1 DS231940 EDS28018.1 AAAB01008847 EGK96605.1 KQ759841 OAD62628.1 AXCM01007783 GBBI01002406 JAC16306.1 AXCN02001373 EDO64304.2 APCN01001005 ADMH02000784 ETN65060.1 UFQS01000248 UFQT01000248 SSX02006.1 SSX22383.1 EGK96606.1 SSX02005.1 SSX22382.1 ACPB03006290 GEZM01063783 JAV68959.1 UFQT01000851 SSX27650.1 GEZM01063784 JAV68958.1 AJWK01020210 APGK01048498 KB741112 KB632184 ENN73817.1 ERL89632.1 LBMM01000050 KMR05159.1 GBHO01039432 GBHO01039429 JAG04172.1 JAG04175.1 CVRI01000063 CRL04860.1 DS235846 EEB18527.1 LNIX01000009 OXA49974.1 JH431897 GL732566 EFX76773.1 OXA49975.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000005204
UP000053240
UP000005203
+ More
UP000076502 UP000078541 UP000053825 UP000007755 UP000279307 UP000005205 UP000078540 UP000008237 UP000078492 UP000192223 UP000007266 UP000037510 UP000235965 UP000008820 UP000079169 UP000069940 UP000249989 UP000030765 UP000002320 UP000007062 UP000069272 UP000075883 UP000076408 UP000075886 UP000075920 UP000075900 UP000075903 UP000075880 UP000075885 UP000075884 UP000075881 UP000075840 UP000000673 UP000075901 UP000076407 UP000075902 UP000015103 UP000092461 UP000019118 UP000030742 UP000036403 UP000183832 UP000009046 UP000198287 UP000000305
UP000076502 UP000078541 UP000053825 UP000007755 UP000279307 UP000005205 UP000078540 UP000008237 UP000078492 UP000192223 UP000007266 UP000037510 UP000235965 UP000008820 UP000079169 UP000069940 UP000249989 UP000030765 UP000002320 UP000007062 UP000069272 UP000075883 UP000076408 UP000075886 UP000075920 UP000075900 UP000075903 UP000075880 UP000075885 UP000075884 UP000075881 UP000075840 UP000000673 UP000075901 UP000076407 UP000075902 UP000015103 UP000092461 UP000019118 UP000030742 UP000036403 UP000183832 UP000009046 UP000198287 UP000000305
PRIDE
Pfam
PF07690 MFS_1
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A2A4IW41
A0A212FAI9
A0A194PL08
H9JPN4
A0A2W1BL11
A0A194RQQ6
+ More
A0A088ACW2 A0A154NYQ3 A0A195FIP1 A0A0L7QPC4 F4WWQ8 A0A3L8DYK7 A0A158NJ06 A0A195BL72 E2BH66 A0A151JN04 A0A1W4X8F4 A0A139WN08 A0A1B6KVF5 A0A0L7LU93 A0A1B6CYN4 A0A1W4X8H0 A0A1W4XIT9 A0A2J7QPW8 Q16M19 A0A1S4EGT1 A0A182GH87 A0A084WPL5 B0WHV8 F5HJZ0 A0A310SIM6 A0A182FJZ9 A0A182M0I4 A0A023F483 A0A182YF02 A0A182Q1B4 A0A182VVB2 A0A182RJ17 A0A182VKW5 A7USK6 A0A182J697 A0A182PM63 A0A182N0Z2 A0A182KFP7 A0A182I9M4 W5JQ81 A0A182SPN8 A0A182X5B1 A0A182TKB4 A0A336KAV3 F5HJY9 A0A336LWF0 T1I7F8 A0A1Y1L5L1 A0A336MCK4 A0A1Y1L844 A0A1B0EVG7 N6U5G7 A0A0J7P5H8 A0A0A9W6V5 A0A1J1IZ92 E0VYS1 A0A226DYV1 T1J6X6 E9GUQ3 A0A226DWW9
A0A088ACW2 A0A154NYQ3 A0A195FIP1 A0A0L7QPC4 F4WWQ8 A0A3L8DYK7 A0A158NJ06 A0A195BL72 E2BH66 A0A151JN04 A0A1W4X8F4 A0A139WN08 A0A1B6KVF5 A0A0L7LU93 A0A1B6CYN4 A0A1W4X8H0 A0A1W4XIT9 A0A2J7QPW8 Q16M19 A0A1S4EGT1 A0A182GH87 A0A084WPL5 B0WHV8 F5HJZ0 A0A310SIM6 A0A182FJZ9 A0A182M0I4 A0A023F483 A0A182YF02 A0A182Q1B4 A0A182VVB2 A0A182RJ17 A0A182VKW5 A7USK6 A0A182J697 A0A182PM63 A0A182N0Z2 A0A182KFP7 A0A182I9M4 W5JQ81 A0A182SPN8 A0A182X5B1 A0A182TKB4 A0A336KAV3 F5HJY9 A0A336LWF0 T1I7F8 A0A1Y1L5L1 A0A336MCK4 A0A1Y1L844 A0A1B0EVG7 N6U5G7 A0A0J7P5H8 A0A0A9W6V5 A0A1J1IZ92 E0VYS1 A0A226DYV1 T1J6X6 E9GUQ3 A0A226DWW9
Ontologies
GO
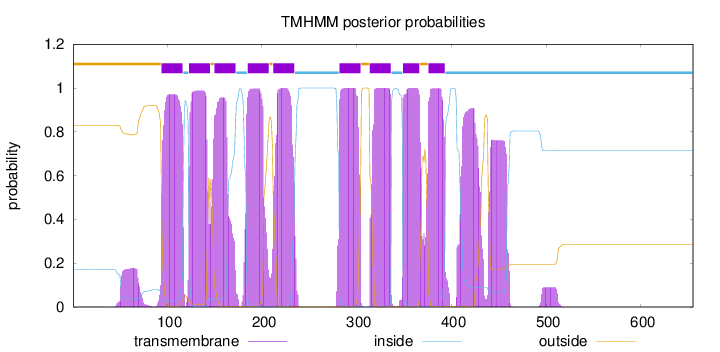
Topology
Length:
655
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
227.69252
Exp number, first 60 AAs:
1.71688
Total prob of N-in:
0.17073
outside
1 - 93
TMhelix
94 - 116
inside
117 - 122
TMhelix
123 - 145
outside
146 - 149
TMhelix
150 - 172
inside
173 - 184
TMhelix
185 - 207
outside
208 - 211
TMhelix
212 - 234
inside
235 - 281
TMhelix
282 - 304
outside
305 - 313
TMhelix
314 - 336
inside
337 - 348
TMhelix
349 - 366
outside
367 - 375
TMhelix
376 - 393
inside
394 - 655
Population Genetic Test Statistics
Pi
254.060373
Theta
184.45818
Tajima's D
1.480838
CLR
0.436378
CSRT
0.782910854457277
Interpretation
Uncertain
