Gene
KWMTBOMO13748
Pre Gene Modal
BGIBMGA011523
Annotation
PREDICTED:_inhibitor_of_apoptosis_2_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.272
Sequence
CDS
ATGAATTATGAGACAAACAGACTCAATACGTTCACAAACTGGCCTGCTTTGGCTCCAGTGGATCCCATCAGAATAGCGAAGGCTGGTTTTTTCTACACTGGCCAAGGAATGGAAGTGCAATGTTTTAGTTGTGGTGGTAAAATTTCCGAGTGGAATTACGGAGATCAAGTTATGTGGCGACATCGTAGGATGGAACCAAATTGTACGTTTGTTGTGAATTCACAACTATCGGGTAATGTACCGATGGTATTAGGCCCTGAATGTCCTTCGACGGATGAGCTAGCCAGGTCTTGGTCGACTACAGATGATCAAAGTGATTTAGTTGAATCTACTGGAGACTATGGTGTTACTGAAGAAGATGAAATGTATAGGAGCGAAGCTTTGCGACTTCTTTCATTTAGTCATTGGGAGGATGATTCTGTATCTCGTGAAGCTCTCGTAAGTGCTGGTTTCTATCACATAGGCGGTGGACGTCTACGATGCGCATGGTGTGGGGGAGAGTTAGCTCCATTCAGACGATTCGGATCATTGGGTCGGCCACTTGAAGTCCATAGAATGTACTTCCCCCGATGCGCCCACGCTGCTGCACTTGAATCGGCACAACAGCCCTTTACTTTACCGACAGTTATGTCGTCACCGTCAAACACGCCCCCAATACAGTCACCACCAGAGACAGGCAATGATTCACAATCACAGTCTGCGGGAGCTGCCCACAACGAGCGACTCCTGAACATGGGCGGTACTTGGCGTACGTTAGGTGTTGTCAGCGGCGGAGCTAGATACCCTCAATACGCGTCACTGGCGTCACGACTGGCGACCTTCGACTCTTGGCCAACCGACAAACAGCAAACGCCTAAAGATCTATCAGAAGCAGGATTCTTTCACACCGGAACTGATGATCAGGTTCGCTGTTTCTACTGTGACGGTGGGCTCGGCAAGTGGGAGGCTGGCGACGCGCCGTGGACGGAGCACGCGCGCTGGTTCCCGCACTGCGGGTACGTGCTGCTCCTCAAGGGAAACGAATTCGTCGATGAGTGCCGAAACGACAATAATCGGAGAAATAATTCACGAGAAACTGGTGTTAGTTCGAGACGAAGAAATCCGGGCGTCAACTACCCGGTGACCGAGATTGAAGTCGAGGAGTGCATGGAGAGTCCAATAACGTTGGCGGCGCTGGGGGCGGGGCTCGACGTCGCGAGGGTGCGCCGAGCTATCCTGCAACGATTGAGATCAACAGGTACACCGTTCCAGACTTCAGAGGCACTGATTGATTCCGTACTAGACGCGCAACTAAACGAGGAAGCTTGGAGTACTAATTCGCAGTCCCAAAGCCTTTCGAGGCATTTGGCTGAAACGCTCAGAGGAATGGATTCAATACCAGGTATAGCACCGCAGTCGTGGGACAACAACCCGGATAGAGGCAGATCGCAAAGAGACGGTATATCTCCGTCGCTTACTGAAATCGAATCGAGGCGGTCTACCAGCCAACCGCGAACATTGCCACCGCGCAGGAACGTATCTGTTATCGATGACGACAGAACTGAGCCTCTGCATAAAGAGGAAAGCTTAACGTTGGAAGAAGAGAATAGACAGCTCAGAGAGGCAAGGCTGTGTAAAGTTTGTATGGATAACGAGGTGAGCGTGGTGTTCCTGCCGTGCGGGCACCTGGTGTCGTGCGCGAGGTGCGGCGCGGCGCTGAGCGCGTGCCCGCTGTGCCGCGGCGCCGTGCGCGCGCTCGTGCGTGCCTACCTCGCCTGA
Protein
MNYETNRLNTFTNWPALAPVDPIRIAKAGFFYTGQGMEVQCFSCGGKISEWNYGDQVMWRHRRMEPNCTFVVNSQLSGNVPMVLGPECPSTDELARSWSTTDDQSDLVESTGDYGVTEEDEMYRSEALRLLSFSHWEDDSVSREALVSAGFYHIGGGRLRCAWCGGELAPFRRFGSLGRPLEVHRMYFPRCAHAAALESAQQPFTLPTVMSSPSNTPPIQSPPETGNDSQSQSAGAAHNERLLNMGGTWRTLGVVSGGARYPQYASLASRLATFDSWPTDKQQTPKDLSEAGFFHTGTDDQVRCFYCDGGLGKWEAGDAPWTEHARWFPHCGYVLLLKGNEFVDECRNDNNRRNNSRETGVSSRRRNPGVNYPVTEIEVEECMESPITLAALGAGLDVARVRRAILQRLRSTGTPFQTSEALIDSVLDAQLNEEAWSTNSQSQSLSRHLAETLRGMDSIPGIAPQSWDNNPDRGRSQRDGISPSLTEIESRRSTSQPRTLPPRRNVSVIDDDRTEPLHKEESLTLEEENRQLREARLCKVCMDNEVSVVFLPCGHLVSCARCGAALSACPLCRGAVRALVRAYLA
Summary
Uniprot
E9JEI0
A0A2H1VF62
A0A2A4JCK1
A0A2H4WW88
A0A194RQQ0
A0A194PMR4
+ More
A0A346RAD8 A0A212FFM8 A0A1B6IHN5 A0A1B6I1E1 A0A1B6FK33 A0A2J7QQ30 A0A2J7QQ42 A0A2J7QQ45 A0A1B6CXZ9 A0A1Y1MZ67 D6WAN9 A0A1W4X4N2 A0A088A7X4 A0A2R7W376 A0A026W8F5 A0A3L8DKD2 A0A0L7RBG0 A0A0A9Y257 A0A146KLT3 A0A232EW65 T1HSP8 K7IZE2 A0A195BRM5 A0A1I8N363 A0A1B0GLP2 A0A195EG15 A0A3Q0IRH0 A0A1L8DJN5 A0A0K2SWB7 A0A076FES9 A0A1A9WAE9 Q2PQQ2 A0A1A9ZRA6 D3TPA6 A0A1A9UWA5 A0A1B0ARP0 B4MQL3 B4QH01 A0A1W4UZ07 A0A1A9YJM6 T1IM88 A0A087UNH8 Q175J8 A0A1S3I9X7 A0A1S4FEB1 B4P6U5 A0A336LPZ5 A0A336LCV0 A0A2L2Y8S3 A0A1S3K6U7 Q28WY9 B4HBE3 B3MC17 A0A2L2Y8S6 Q5XIW4 F7FLN8 A0A1E1X889 W5JTH2 A0A1S3JEM2 Q9ESE9 A0A1E1XK72 A0A1S3IP31 A0A1S3H002 B0XJ37 A0A1Q3FHI1 M3Y735 V9KD44 L7M5L9 A0A0P8XPZ5 A0A1Q3FHJ8 A0A1Q3FHQ7 A0A131YI84 A0A3P9PTC7 A0A084WCJ9 A0A1I8JSI2 A0A0S7K059 A0A3B5MI01 A0A1S3HWJ8 A0A1A7WMY4 G3SXE7 M4A8R3 A0A3Q7TMM9 A0A2I4AT26 A0A1A8FUI9 A0A1A8M9H7 A0A3Q1JK44 A0A087YKG0 A0A3B3U0R1 A0A1A8SC81 A0A1A8I9Z8
A0A346RAD8 A0A212FFM8 A0A1B6IHN5 A0A1B6I1E1 A0A1B6FK33 A0A2J7QQ30 A0A2J7QQ42 A0A2J7QQ45 A0A1B6CXZ9 A0A1Y1MZ67 D6WAN9 A0A1W4X4N2 A0A088A7X4 A0A2R7W376 A0A026W8F5 A0A3L8DKD2 A0A0L7RBG0 A0A0A9Y257 A0A146KLT3 A0A232EW65 T1HSP8 K7IZE2 A0A195BRM5 A0A1I8N363 A0A1B0GLP2 A0A195EG15 A0A3Q0IRH0 A0A1L8DJN5 A0A0K2SWB7 A0A076FES9 A0A1A9WAE9 Q2PQQ2 A0A1A9ZRA6 D3TPA6 A0A1A9UWA5 A0A1B0ARP0 B4MQL3 B4QH01 A0A1W4UZ07 A0A1A9YJM6 T1IM88 A0A087UNH8 Q175J8 A0A1S3I9X7 A0A1S4FEB1 B4P6U5 A0A336LPZ5 A0A336LCV0 A0A2L2Y8S3 A0A1S3K6U7 Q28WY9 B4HBE3 B3MC17 A0A2L2Y8S6 Q5XIW4 F7FLN8 A0A1E1X889 W5JTH2 A0A1S3JEM2 Q9ESE9 A0A1E1XK72 A0A1S3IP31 A0A1S3H002 B0XJ37 A0A1Q3FHI1 M3Y735 V9KD44 L7M5L9 A0A0P8XPZ5 A0A1Q3FHJ8 A0A1Q3FHQ7 A0A131YI84 A0A3P9PTC7 A0A084WCJ9 A0A1I8JSI2 A0A0S7K059 A0A3B5MI01 A0A1S3HWJ8 A0A1A7WMY4 G3SXE7 M4A8R3 A0A3Q7TMM9 A0A2I4AT26 A0A1A8FUI9 A0A1A8M9H7 A0A3Q1JK44 A0A087YKG0 A0A3B3U0R1 A0A1A8SC81 A0A1A8I9Z8
Pubmed
19121390
21040523
26354079
29910977
22118469
28004739
+ More
18362917 19820115 24508170 30249741 25401762 26823975 28648823 20075255 25315136 16907828 20353571 17994087 22936249 17510324 17550304 26561354 15632085 15489334 15057822 15632090 28503490 20920257 23761445 11860601 29209593 24402279 25576852 26830274 24438588 23542700
18362917 19820115 24508170 30249741 25401762 26823975 28648823 20075255 25315136 16907828 20353571 17994087 22936249 17510324 17550304 26561354 15632085 15489334 15057822 15632090 28503490 20920257 23761445 11860601 29209593 24402279 25576852 26830274 24438588 23542700
EMBL
BABH01015194
BABH01015195
BABH01015196
GQ426297
ADM32523.1
ODYU01002254
+ More
SOQ39500.1 NWSH01002058 PCG69284.1 KY514096 AUD12127.1 KQ459833 KPJ19812.1 KQ459601 KPI94024.1 MH200928 AXS59138.1 AGBW02008806 OWR52523.1 GECU01021262 JAS86444.1 GECU01026973 JAS80733.1 GECZ01019224 JAS50545.1 NEVH01012087 PNF30694.1 PNF30691.1 PNF30693.1 GEDC01018949 JAS18349.1 GEZM01020453 JAV89266.1 JN108872 KQ971312 AEQ93553.1 EEZ99286.1 KK854230 PTY13440.1 KK107347 EZA52248.1 QOIP01000007 RLU20359.1 KQ414617 KOC68081.1 GBHO01016502 GBRD01015606 JAG27102.1 JAG50220.1 GDHC01021118 JAP97510.1 NNAY01001909 OXU22592.1 ACPB03006767 KQ976424 KYM88604.1 AJWK01002077 KQ978983 KYN26817.1 GFDF01007524 JAV06560.1 HACA01000444 CDW17805.1 KF516634 AII16538.1 DQ307171 ABC25071.1 CCAG010009975 EZ423258 ADD19534.1 JXJN01002494 CH963849 EDW74402.1 CM000362 CM002911 EDX07264.1 KMY94105.1 JH430992 KK120738 KFM78917.1 CH477399 EAT41756.2 CM000158 EDW92022.1 UFQS01000042 UFQT01000042 SSW98317.1 SSX18703.1 UFQS01002793 UFQT01002793 SSX14681.1 SSX34078.1 IAAA01018954 LAA04566.1 CM000071 EAL26527.1 CH479252 EDW38685.1 CH902619 EDV37204.1 IAAA01018953 LAA04562.1 AABR07069023 BC083555 CH473993 AAH83555.1 EDL78521.1 GFAC01003728 JAT95460.1 ADMH02000540 ETN66039.1 AF183430 AAG22970.1 GFAA01003717 JAT99717.1 DS233428 EDS29932.1 GFDL01008037 JAV27008.1 AEYP01061514 JW863297 AFO95814.1 GACK01005887 JAA59147.1 KPU76649.1 GFDL01008005 JAV27040.1 GFDL01007944 JAV27101.1 GEDV01010312 JAP78245.1 ATLV01022681 KE525335 KFB47943.1 GBYX01225374 GBYX01225372 JAO70434.1 HADW01005723 HADX01009860 SBP07123.1 HAEB01015257 HAEC01002013 SBQ61784.1 HAEF01011795 HAEG01010786 SBR52954.1 AYCK01000596 HAEH01014207 HAEI01013388 SBS15857.1 HAED01007715 HAEE01013630 SBQ93927.1
SOQ39500.1 NWSH01002058 PCG69284.1 KY514096 AUD12127.1 KQ459833 KPJ19812.1 KQ459601 KPI94024.1 MH200928 AXS59138.1 AGBW02008806 OWR52523.1 GECU01021262 JAS86444.1 GECU01026973 JAS80733.1 GECZ01019224 JAS50545.1 NEVH01012087 PNF30694.1 PNF30691.1 PNF30693.1 GEDC01018949 JAS18349.1 GEZM01020453 JAV89266.1 JN108872 KQ971312 AEQ93553.1 EEZ99286.1 KK854230 PTY13440.1 KK107347 EZA52248.1 QOIP01000007 RLU20359.1 KQ414617 KOC68081.1 GBHO01016502 GBRD01015606 JAG27102.1 JAG50220.1 GDHC01021118 JAP97510.1 NNAY01001909 OXU22592.1 ACPB03006767 KQ976424 KYM88604.1 AJWK01002077 KQ978983 KYN26817.1 GFDF01007524 JAV06560.1 HACA01000444 CDW17805.1 KF516634 AII16538.1 DQ307171 ABC25071.1 CCAG010009975 EZ423258 ADD19534.1 JXJN01002494 CH963849 EDW74402.1 CM000362 CM002911 EDX07264.1 KMY94105.1 JH430992 KK120738 KFM78917.1 CH477399 EAT41756.2 CM000158 EDW92022.1 UFQS01000042 UFQT01000042 SSW98317.1 SSX18703.1 UFQS01002793 UFQT01002793 SSX14681.1 SSX34078.1 IAAA01018954 LAA04566.1 CM000071 EAL26527.1 CH479252 EDW38685.1 CH902619 EDV37204.1 IAAA01018953 LAA04562.1 AABR07069023 BC083555 CH473993 AAH83555.1 EDL78521.1 GFAC01003728 JAT95460.1 ADMH02000540 ETN66039.1 AF183430 AAG22970.1 GFAA01003717 JAT99717.1 DS233428 EDS29932.1 GFDL01008037 JAV27008.1 AEYP01061514 JW863297 AFO95814.1 GACK01005887 JAA59147.1 KPU76649.1 GFDL01008005 JAV27040.1 GFDL01007944 JAV27101.1 GEDV01010312 JAP78245.1 ATLV01022681 KE525335 KFB47943.1 GBYX01225374 GBYX01225372 JAO70434.1 HADW01005723 HADX01009860 SBP07123.1 HAEB01015257 HAEC01002013 SBQ61784.1 HAEF01011795 HAEG01010786 SBR52954.1 AYCK01000596 HAEH01014207 HAEI01013388 SBS15857.1 HAED01007715 HAEE01013630 SBQ93927.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000235965
+ More
UP000007266 UP000192223 UP000005203 UP000053097 UP000279307 UP000053825 UP000215335 UP000015103 UP000002358 UP000078540 UP000095301 UP000092461 UP000078492 UP000079169 UP000091820 UP000092445 UP000092444 UP000078200 UP000092460 UP000007798 UP000000304 UP000192221 UP000092443 UP000054359 UP000008820 UP000085678 UP000002282 UP000001819 UP000008744 UP000007801 UP000002494 UP000000673 UP000002320 UP000000715 UP000242638 UP000030765 UP000069272 UP000261380 UP000007646 UP000002852 UP000286640 UP000192220 UP000265040 UP000028760 UP000261500
UP000007266 UP000192223 UP000005203 UP000053097 UP000279307 UP000053825 UP000215335 UP000015103 UP000002358 UP000078540 UP000095301 UP000092461 UP000078492 UP000079169 UP000091820 UP000092445 UP000092444 UP000078200 UP000092460 UP000007798 UP000000304 UP000192221 UP000092443 UP000054359 UP000008820 UP000085678 UP000002282 UP000001819 UP000008744 UP000007801 UP000002494 UP000000673 UP000002320 UP000000715 UP000242638 UP000030765 UP000069272 UP000261380 UP000007646 UP000002852 UP000286640 UP000192220 UP000265040 UP000028760 UP000261500
Interpro
SUPFAM
SSF47986
SSF47986
Gene 3D
ProteinModelPortal
E9JEI0
A0A2H1VF62
A0A2A4JCK1
A0A2H4WW88
A0A194RQQ0
A0A194PMR4
+ More
A0A346RAD8 A0A212FFM8 A0A1B6IHN5 A0A1B6I1E1 A0A1B6FK33 A0A2J7QQ30 A0A2J7QQ42 A0A2J7QQ45 A0A1B6CXZ9 A0A1Y1MZ67 D6WAN9 A0A1W4X4N2 A0A088A7X4 A0A2R7W376 A0A026W8F5 A0A3L8DKD2 A0A0L7RBG0 A0A0A9Y257 A0A146KLT3 A0A232EW65 T1HSP8 K7IZE2 A0A195BRM5 A0A1I8N363 A0A1B0GLP2 A0A195EG15 A0A3Q0IRH0 A0A1L8DJN5 A0A0K2SWB7 A0A076FES9 A0A1A9WAE9 Q2PQQ2 A0A1A9ZRA6 D3TPA6 A0A1A9UWA5 A0A1B0ARP0 B4MQL3 B4QH01 A0A1W4UZ07 A0A1A9YJM6 T1IM88 A0A087UNH8 Q175J8 A0A1S3I9X7 A0A1S4FEB1 B4P6U5 A0A336LPZ5 A0A336LCV0 A0A2L2Y8S3 A0A1S3K6U7 Q28WY9 B4HBE3 B3MC17 A0A2L2Y8S6 Q5XIW4 F7FLN8 A0A1E1X889 W5JTH2 A0A1S3JEM2 Q9ESE9 A0A1E1XK72 A0A1S3IP31 A0A1S3H002 B0XJ37 A0A1Q3FHI1 M3Y735 V9KD44 L7M5L9 A0A0P8XPZ5 A0A1Q3FHJ8 A0A1Q3FHQ7 A0A131YI84 A0A3P9PTC7 A0A084WCJ9 A0A1I8JSI2 A0A0S7K059 A0A3B5MI01 A0A1S3HWJ8 A0A1A7WMY4 G3SXE7 M4A8R3 A0A3Q7TMM9 A0A2I4AT26 A0A1A8FUI9 A0A1A8M9H7 A0A3Q1JK44 A0A087YKG0 A0A3B3U0R1 A0A1A8SC81 A0A1A8I9Z8
A0A346RAD8 A0A212FFM8 A0A1B6IHN5 A0A1B6I1E1 A0A1B6FK33 A0A2J7QQ30 A0A2J7QQ42 A0A2J7QQ45 A0A1B6CXZ9 A0A1Y1MZ67 D6WAN9 A0A1W4X4N2 A0A088A7X4 A0A2R7W376 A0A026W8F5 A0A3L8DKD2 A0A0L7RBG0 A0A0A9Y257 A0A146KLT3 A0A232EW65 T1HSP8 K7IZE2 A0A195BRM5 A0A1I8N363 A0A1B0GLP2 A0A195EG15 A0A3Q0IRH0 A0A1L8DJN5 A0A0K2SWB7 A0A076FES9 A0A1A9WAE9 Q2PQQ2 A0A1A9ZRA6 D3TPA6 A0A1A9UWA5 A0A1B0ARP0 B4MQL3 B4QH01 A0A1W4UZ07 A0A1A9YJM6 T1IM88 A0A087UNH8 Q175J8 A0A1S3I9X7 A0A1S4FEB1 B4P6U5 A0A336LPZ5 A0A336LCV0 A0A2L2Y8S3 A0A1S3K6U7 Q28WY9 B4HBE3 B3MC17 A0A2L2Y8S6 Q5XIW4 F7FLN8 A0A1E1X889 W5JTH2 A0A1S3JEM2 Q9ESE9 A0A1E1XK72 A0A1S3IP31 A0A1S3H002 B0XJ37 A0A1Q3FHI1 M3Y735 V9KD44 L7M5L9 A0A0P8XPZ5 A0A1Q3FHJ8 A0A1Q3FHQ7 A0A131YI84 A0A3P9PTC7 A0A084WCJ9 A0A1I8JSI2 A0A0S7K059 A0A3B5MI01 A0A1S3HWJ8 A0A1A7WMY4 G3SXE7 M4A8R3 A0A3Q7TMM9 A0A2I4AT26 A0A1A8FUI9 A0A1A8M9H7 A0A3Q1JK44 A0A087YKG0 A0A3B3U0R1 A0A1A8SC81 A0A1A8I9Z8
PDB
3T6P
E-value=4.6949e-47,
Score=475
Ontologies
PATHWAY
GO
GO:1990001
GO:0031398
GO:0061630
GO:0043154
GO:0005634
GO:0043027
GO:0005737
GO:0016021
GO:0048471
GO:0006964
GO:0061057
GO:0006511
GO:0089720
GO:0070534
GO:0031625
GO:0050829
GO:0070936
GO:0022416
GO:0051291
GO:0007283
GO:0032991
GO:0045121
GO:0060546
GO:0043066
GO:0005654
GO:0005829
GO:0042981
GO:0060544
GO:0004842
GO:0005515
GO:0008270
GO:0000287
GO:0016620
GO:0016787
GO:0003676
GO:0016491
GO:0015930
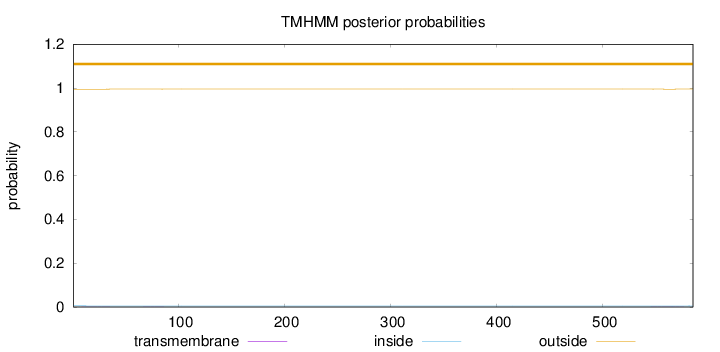
Topology
Length:
585
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0418599999999999
Exp number, first 60 AAs:
0.01439
Total prob of N-in:
0.00542
outside
1 - 585
Population Genetic Test Statistics
Pi
203.538238
Theta
194.702581
Tajima's D
-1.630745
CLR
14.872814
CSRT
0.0443477826108695
Interpretation
Possibly Positive selection
