Pre Gene Modal
BGIBMGA011491
Annotation
PREDICTED:_3-ketodihydrosphingosine_reductase_[Amyelois_transitella]
Full name
3-ketodihydrosphingosine reductase
Alternative Name
3-dehydrosphinganine reductase
Follicular variant translocation protein 1
Short chain dehydrogenase/reductase family 35C member 1
Follicular variant translocation protein 1
Short chain dehydrogenase/reductase family 35C member 1
Location in the cell
PlasmaMembrane Reliability : 3.75
Sequence
CDS
ATGTTGCTAACAGTTGGAATTTTAATAACATTTTTAGTACTGGTCTTATCATTAGTTTTATATTTAACGATAGAAAAAAAAGTTAGTTACAAGGACTTGTTGAATAAACATGTTCTAATTACCGGTGGATCTAGTGGAATTGGAAAATCCGCTGCTATTGAAGCTGCAAGACGTGGAGCTCATGTCTCTATCATCGGTCGAGACGAAAATAAACTTAAGTCAGCTGTGGAAGAAATAACTTCACAGTGTTTAGATAAGTCTGTTCAATCCATTCAATATGCAGTTCTGGATGTTACATCTGACTATTTCGTTATAGAAAAAGCGATAAATTCGTTAGAGGACAATGTAGGACCAATATTTATGTTGGTAAACTGCGCCGGCATGTGTATATGTGGTAAATTTGAACAAATGAAGGTAGAGGATATAAAACAGATGATTGACTTGAATTATTTTGGCAGTGCGTATATGACAAAATGTGTTTTGCCGGGCATGAAGAAAAAAAAAGAAGGATTGATCGTATTTGTATGTTCAGAAGCAGCTTTAATAGGCATTTATGGATACACAGCATACAGTGCTGGAAAATGGGCTGTGCGAGGTTTAGCTGAGGCCCTGAGTATGGAGTTAATTGGAACTGGTGTCCGAGTAATGATATCATATCCTCCTGATACAGACACCCCGGGTTTTAAGAATGAAGATCTTACAAAGCCTTTAGAAACCAAACTTATATCAGGCTCTGCTGGTCTACTTTCTCCAGATGTAGTTGGAAAGCAAATGATAGATGATGCATTGAATGGGAAAATGTATTCCGTGTATACTACATCTGGATCAATGTTAACAACACTGTTTGGTGGTTCAATTGATAGTGTACGTCAAATTTTACTACAAGTGTTTTCAATGGGAGTTTTACGTGCAGTTATGGTAGGGGTTCTATTGTCATTTCATAAAATTGTGCGAGACAATTTAAAGTTGCAAAACAATAGCAAAAACAAATAA
Protein
MLLTVGILITFLVLVLSLVLYLTIEKKVSYKDLLNKHVLITGGSSGIGKSAAIEAARRGAHVSIIGRDENKLKSAVEEITSQCLDKSVQSIQYAVLDVTSDYFVIEKAINSLEDNVGPIFMLVNCAGMCICGKFEQMKVEDIKQMIDLNYFGSAYMTKCVLPGMKKKKEGLIVFVCSEAALIGIYGYTAYSAGKWAVRGLAEALSMELIGTGVRVMISYPPDTDTPGFKNEDLTKPLETKLISGSAGLLSPDVVGKQMIDDALNGKMYSVYTTSGSMLTTLFGGSIDSVRQILLQVFSMGVLRAVMVGVLLSFHKIVRDNLKLQNNSKNK
Summary
Description
Catalyzes the reduction of 3-ketodihydrosphingosine (KDS) to dihydrosphingosine (DHS).
Catalytic Activity
NADP(+) + sphinganine = 3-oxosphinganine + H(+) + NADPH
Similarity
Belongs to the short-chain dehydrogenases/reductases (SDR) family.
Keywords
Alternative splicing
Chromosomal rearrangement
Complete proteome
Disease mutation
Endoplasmic reticulum
Lipid metabolism
Membrane
NADP
Oxidoreductase
Palmoplantar keratoderma
Proto-oncogene
Reference proteome
Signal
Sphingolipid metabolism
Transmembrane
Transmembrane helix
Feature
chain 3-ketodihydrosphingosine reductase
splice variant In isoform 2.
sequence variant In EKVP4; loss of 3-dehydrosphinganine reductase activity.
splice variant In isoform 2.
sequence variant In EKVP4; loss of 3-dehydrosphinganine reductase activity.
Uniprot
H9JPN6
A0A2A4JB29
A0A2W1B0T9
A0A2H1VF95
A0A3S2MA56
A0A194RUJ6
+ More
S4PH41 A0A212FFL4 A0A2J7R6X5 A0A067RUJ4 A0A0L7RFC7 A0A2P8XNT4 A0A1B6FIX8 A0A154PFB3 A0A1B6HYQ1 A0A087ZRV0 A0A2J7R6U9 T1J3U3 A0A026WWB1 A0A2A3EP45 A0A023F2N5 A0A0C9QE85 A0A1B6CKZ1 A0A310SBW5 A0A1Y1M237 A0A1L8E9M7 A0A0P4VRF5 A0A224XT62 A0A1L8ED16 A0A195DZY6 A0A1S4EXM3 Q17MF6 E2AQ98 V4AMH6 A0A195CR02 A0A1I8P0X5 A0A182GB89 A0A182N6G6 A0A3Q3K1S5 A0A182SW23 A0A182Q769 T1HQ05 E9IJB7 A0A195BWK1 A0A182XXP9 A0A158NEN3 A0A182XH49 F4WRR8 A0A151XJ90 E2B3B7 A0A182VQJ8 Q7Q282 A0A1I8N6V1 A0A182I8A3 A0A182KMR1 A0A182U3W7 A0A182V4V7 A0A2S2QUB2 D6W737 A0A195FKP8 A0A3P9N9B9 A0A087XH72 A0A3B4A3H2 A0A3B3UBU8 A0A3B3Y9H1 A0A2R9ASL0 A0A182RQR8 K7IVW1 H2MQK7 A0A2I4D1Z1 A0A2J8W3J5 H2QEM9 A0A024R292 Q06136 G3RBX4 M4APX0 A0A3B5M804 B4GEZ7 Q296L7 A0A3Q2PPP7 A0A182PF25 A0A182M1V8 A0A3Q3AMH0 A0A146NWA9 A0A3P9KR20 A0A2K6SUY2 G2HHD2 F7GGW4 S7Q8Y4 A0A0A9YHD7 A0A182JZT0 A0A182FL40 I1SRY0 A0A3B3BCX7 A0A3Q1I1S9 Q803B4 A0A3B5BAG1
S4PH41 A0A212FFL4 A0A2J7R6X5 A0A067RUJ4 A0A0L7RFC7 A0A2P8XNT4 A0A1B6FIX8 A0A154PFB3 A0A1B6HYQ1 A0A087ZRV0 A0A2J7R6U9 T1J3U3 A0A026WWB1 A0A2A3EP45 A0A023F2N5 A0A0C9QE85 A0A1B6CKZ1 A0A310SBW5 A0A1Y1M237 A0A1L8E9M7 A0A0P4VRF5 A0A224XT62 A0A1L8ED16 A0A195DZY6 A0A1S4EXM3 Q17MF6 E2AQ98 V4AMH6 A0A195CR02 A0A1I8P0X5 A0A182GB89 A0A182N6G6 A0A3Q3K1S5 A0A182SW23 A0A182Q769 T1HQ05 E9IJB7 A0A195BWK1 A0A182XXP9 A0A158NEN3 A0A182XH49 F4WRR8 A0A151XJ90 E2B3B7 A0A182VQJ8 Q7Q282 A0A1I8N6V1 A0A182I8A3 A0A182KMR1 A0A182U3W7 A0A182V4V7 A0A2S2QUB2 D6W737 A0A195FKP8 A0A3P9N9B9 A0A087XH72 A0A3B4A3H2 A0A3B3UBU8 A0A3B3Y9H1 A0A2R9ASL0 A0A182RQR8 K7IVW1 H2MQK7 A0A2I4D1Z1 A0A2J8W3J5 H2QEM9 A0A024R292 Q06136 G3RBX4 M4APX0 A0A3B5M804 B4GEZ7 Q296L7 A0A3Q2PPP7 A0A182PF25 A0A182M1V8 A0A3Q3AMH0 A0A146NWA9 A0A3P9KR20 A0A2K6SUY2 G2HHD2 F7GGW4 S7Q8Y4 A0A0A9YHD7 A0A182JZT0 A0A182FL40 I1SRY0 A0A3B3BCX7 A0A3Q1I1S9 Q803B4 A0A3B5BAG1
EC Number
1.1.1.102
Pubmed
19121390
28756777
26354079
23622113
22118469
24845553
+ More
29403074 24508170 30249741 25474469 28004739 27129103 17510324 20798317 23254933 26483478 21282665 25244985 21347285 21719571 12364791 14747013 17210077 25315136 20966253 18362917 19820115 25463417 22722832 20075255 17554307 16136131 11181995 8417785 14702039 16177791 15489334 15328338 28575652 22398555 23542700 17994087 15632085 21484476 17495919 25401762 22445008 29451363
29403074 24508170 30249741 25474469 28004739 27129103 17510324 20798317 23254933 26483478 21282665 25244985 21347285 21719571 12364791 14747013 17210077 25315136 20966253 18362917 19820115 25463417 22722832 20075255 17554307 16136131 11181995 8417785 14702039 16177791 15489334 15328338 28575652 22398555 23542700 17994087 15632085 21484476 17495919 25401762 22445008 29451363
EMBL
BABH01015193
BABH01015194
NWSH01002058
PCG69285.1
KZ150490
PZC70692.1
+ More
ODYU01002254 SOQ39499.1 RSAL01000002 RVE54926.1 KQ459833 KPJ19811.1 GAIX01003402 JAA89158.1 AGBW02008806 OWR52522.1 NEVH01006750 PNF36566.1 KK852409 KDR24480.1 KQ414606 KOC69550.1 PYGN01001631 PSN33671.1 GECZ01019620 JAS50149.1 KQ434886 KZC10134.1 GECU01027914 JAS79792.1 PNF36564.1 JH431831 KK107078 QOIP01000011 EZA60332.1 RLU16485.1 KZ288202 PBC33470.1 GBBI01003413 JAC15299.1 GBYB01001614 JAG71381.1 GEDC01023263 JAS14035.1 KQ764228 OAD54575.1 GEZM01042532 GEZM01042531 JAV79793.1 GFDG01003361 JAV15438.1 GDKW01001601 JAI54994.1 GFTR01005275 JAW11151.1 GFDG01002274 JAV16525.1 KQ979999 KYN18267.1 CH477206 EAT47882.1 GL441701 EFN64432.1 KB201262 ESO98337.1 KQ977381 KYN03131.1 JXUM01157742 KQ572838 KXJ67985.1 AXCN02000286 ACPB03017380 GL763764 EFZ19326.1 KQ976401 KYM92341.1 ADTU01013565 GL888292 EGI63075.1 KQ982080 KYQ60280.1 GL445323 EFN89795.1 AAAB01008978 EAA13630.3 APCN01002684 GGMS01012122 MBY81325.1 KQ971307 EFA11415.2 KQ981490 KYN41240.1 AYCK01007797 AJFE02095804 AJFE02095805 AJFE02095806 AJFE02095807 AJFE02095808 ABGA01233288 ABGA01233289 ABGA01233290 ABGA01233291 ABGA01233292 ABGA01233293 ABGA01233294 ABGA01233295 NDHI03003401 PNJ64344.1 PNJ64347.1 AACZ04062422 GABC01005629 GABC01005628 GABF01002843 GABF01002842 GABF01002841 GABF01002840 GABD01007742 GABD01007741 GABD01007740 GABD01007739 GABD01007738 GABE01005347 GABE01005346 GABE01005345 GABE01005344 GABE01005343 GABE01005342 NBAG03000231 JAA05709.1 JAA19302.1 JAA25358.1 JAA39392.1 PNI69729.1 CH471096 EAW63141.1 X63657 BT006782 AK297670 AK312360 AC021803 AC036176 BC008797 CABD030109886 CABD030109887 CABD030109888 CH479182 EDW34182.1 CM000070 EAL28440.1 AXCM01002920 GCES01150575 JAQ35747.1 AK306146 BAK63140.1 KE164355 EPQ17382.1 GBHO01011117 GBRD01016859 JAG32487.1 JAG48968.1 HM137285 AEA51181.1 BC044552 AAH44552.1
ODYU01002254 SOQ39499.1 RSAL01000002 RVE54926.1 KQ459833 KPJ19811.1 GAIX01003402 JAA89158.1 AGBW02008806 OWR52522.1 NEVH01006750 PNF36566.1 KK852409 KDR24480.1 KQ414606 KOC69550.1 PYGN01001631 PSN33671.1 GECZ01019620 JAS50149.1 KQ434886 KZC10134.1 GECU01027914 JAS79792.1 PNF36564.1 JH431831 KK107078 QOIP01000011 EZA60332.1 RLU16485.1 KZ288202 PBC33470.1 GBBI01003413 JAC15299.1 GBYB01001614 JAG71381.1 GEDC01023263 JAS14035.1 KQ764228 OAD54575.1 GEZM01042532 GEZM01042531 JAV79793.1 GFDG01003361 JAV15438.1 GDKW01001601 JAI54994.1 GFTR01005275 JAW11151.1 GFDG01002274 JAV16525.1 KQ979999 KYN18267.1 CH477206 EAT47882.1 GL441701 EFN64432.1 KB201262 ESO98337.1 KQ977381 KYN03131.1 JXUM01157742 KQ572838 KXJ67985.1 AXCN02000286 ACPB03017380 GL763764 EFZ19326.1 KQ976401 KYM92341.1 ADTU01013565 GL888292 EGI63075.1 KQ982080 KYQ60280.1 GL445323 EFN89795.1 AAAB01008978 EAA13630.3 APCN01002684 GGMS01012122 MBY81325.1 KQ971307 EFA11415.2 KQ981490 KYN41240.1 AYCK01007797 AJFE02095804 AJFE02095805 AJFE02095806 AJFE02095807 AJFE02095808 ABGA01233288 ABGA01233289 ABGA01233290 ABGA01233291 ABGA01233292 ABGA01233293 ABGA01233294 ABGA01233295 NDHI03003401 PNJ64344.1 PNJ64347.1 AACZ04062422 GABC01005629 GABC01005628 GABF01002843 GABF01002842 GABF01002841 GABF01002840 GABD01007742 GABD01007741 GABD01007740 GABD01007739 GABD01007738 GABE01005347 GABE01005346 GABE01005345 GABE01005344 GABE01005343 GABE01005342 NBAG03000231 JAA05709.1 JAA19302.1 JAA25358.1 JAA39392.1 PNI69729.1 CH471096 EAW63141.1 X63657 BT006782 AK297670 AK312360 AC021803 AC036176 BC008797 CABD030109886 CABD030109887 CABD030109888 CH479182 EDW34182.1 CM000070 EAL28440.1 AXCM01002920 GCES01150575 JAQ35747.1 AK306146 BAK63140.1 KE164355 EPQ17382.1 GBHO01011117 GBRD01016859 JAG32487.1 JAG48968.1 HM137285 AEA51181.1 BC044552 AAH44552.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000007151
UP000235965
+ More
UP000027135 UP000053825 UP000245037 UP000076502 UP000005203 UP000053097 UP000279307 UP000242457 UP000078492 UP000008820 UP000000311 UP000030746 UP000078542 UP000095300 UP000069940 UP000249989 UP000075884 UP000261600 UP000075901 UP000075886 UP000015103 UP000078540 UP000076408 UP000005205 UP000076407 UP000007755 UP000075809 UP000008237 UP000075920 UP000007062 UP000095301 UP000075840 UP000075882 UP000075902 UP000075903 UP000007266 UP000078541 UP000242638 UP000028760 UP000261520 UP000261500 UP000261480 UP000240080 UP000075900 UP000002358 UP000001038 UP000192220 UP000001595 UP000002277 UP000005640 UP000001519 UP000002852 UP000261380 UP000008744 UP000001819 UP000265000 UP000075885 UP000075883 UP000264800 UP000265180 UP000233220 UP000002280 UP000075881 UP000069272 UP000261560 UP000257200 UP000261400
UP000027135 UP000053825 UP000245037 UP000076502 UP000005203 UP000053097 UP000279307 UP000242457 UP000078492 UP000008820 UP000000311 UP000030746 UP000078542 UP000095300 UP000069940 UP000249989 UP000075884 UP000261600 UP000075901 UP000075886 UP000015103 UP000078540 UP000076408 UP000005205 UP000076407 UP000007755 UP000075809 UP000008237 UP000075920 UP000007062 UP000095301 UP000075840 UP000075882 UP000075902 UP000075903 UP000007266 UP000078541 UP000242638 UP000028760 UP000261520 UP000261500 UP000261480 UP000240080 UP000075900 UP000002358 UP000001038 UP000192220 UP000001595 UP000002277 UP000005640 UP000001519 UP000002852 UP000261380 UP000008744 UP000001819 UP000265000 UP000075885 UP000075883 UP000264800 UP000265180 UP000233220 UP000002280 UP000075881 UP000069272 UP000261560 UP000257200 UP000261400
PRIDE
Pfam
PF00106 adh_short
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
H9JPN6
A0A2A4JB29
A0A2W1B0T9
A0A2H1VF95
A0A3S2MA56
A0A194RUJ6
+ More
S4PH41 A0A212FFL4 A0A2J7R6X5 A0A067RUJ4 A0A0L7RFC7 A0A2P8XNT4 A0A1B6FIX8 A0A154PFB3 A0A1B6HYQ1 A0A087ZRV0 A0A2J7R6U9 T1J3U3 A0A026WWB1 A0A2A3EP45 A0A023F2N5 A0A0C9QE85 A0A1B6CKZ1 A0A310SBW5 A0A1Y1M237 A0A1L8E9M7 A0A0P4VRF5 A0A224XT62 A0A1L8ED16 A0A195DZY6 A0A1S4EXM3 Q17MF6 E2AQ98 V4AMH6 A0A195CR02 A0A1I8P0X5 A0A182GB89 A0A182N6G6 A0A3Q3K1S5 A0A182SW23 A0A182Q769 T1HQ05 E9IJB7 A0A195BWK1 A0A182XXP9 A0A158NEN3 A0A182XH49 F4WRR8 A0A151XJ90 E2B3B7 A0A182VQJ8 Q7Q282 A0A1I8N6V1 A0A182I8A3 A0A182KMR1 A0A182U3W7 A0A182V4V7 A0A2S2QUB2 D6W737 A0A195FKP8 A0A3P9N9B9 A0A087XH72 A0A3B4A3H2 A0A3B3UBU8 A0A3B3Y9H1 A0A2R9ASL0 A0A182RQR8 K7IVW1 H2MQK7 A0A2I4D1Z1 A0A2J8W3J5 H2QEM9 A0A024R292 Q06136 G3RBX4 M4APX0 A0A3B5M804 B4GEZ7 Q296L7 A0A3Q2PPP7 A0A182PF25 A0A182M1V8 A0A3Q3AMH0 A0A146NWA9 A0A3P9KR20 A0A2K6SUY2 G2HHD2 F7GGW4 S7Q8Y4 A0A0A9YHD7 A0A182JZT0 A0A182FL40 I1SRY0 A0A3B3BCX7 A0A3Q1I1S9 Q803B4 A0A3B5BAG1
S4PH41 A0A212FFL4 A0A2J7R6X5 A0A067RUJ4 A0A0L7RFC7 A0A2P8XNT4 A0A1B6FIX8 A0A154PFB3 A0A1B6HYQ1 A0A087ZRV0 A0A2J7R6U9 T1J3U3 A0A026WWB1 A0A2A3EP45 A0A023F2N5 A0A0C9QE85 A0A1B6CKZ1 A0A310SBW5 A0A1Y1M237 A0A1L8E9M7 A0A0P4VRF5 A0A224XT62 A0A1L8ED16 A0A195DZY6 A0A1S4EXM3 Q17MF6 E2AQ98 V4AMH6 A0A195CR02 A0A1I8P0X5 A0A182GB89 A0A182N6G6 A0A3Q3K1S5 A0A182SW23 A0A182Q769 T1HQ05 E9IJB7 A0A195BWK1 A0A182XXP9 A0A158NEN3 A0A182XH49 F4WRR8 A0A151XJ90 E2B3B7 A0A182VQJ8 Q7Q282 A0A1I8N6V1 A0A182I8A3 A0A182KMR1 A0A182U3W7 A0A182V4V7 A0A2S2QUB2 D6W737 A0A195FKP8 A0A3P9N9B9 A0A087XH72 A0A3B4A3H2 A0A3B3UBU8 A0A3B3Y9H1 A0A2R9ASL0 A0A182RQR8 K7IVW1 H2MQK7 A0A2I4D1Z1 A0A2J8W3J5 H2QEM9 A0A024R292 Q06136 G3RBX4 M4APX0 A0A3B5M804 B4GEZ7 Q296L7 A0A3Q2PPP7 A0A182PF25 A0A182M1V8 A0A3Q3AMH0 A0A146NWA9 A0A3P9KR20 A0A2K6SUY2 G2HHD2 F7GGW4 S7Q8Y4 A0A0A9YHD7 A0A182JZT0 A0A182FL40 I1SRY0 A0A3B3BCX7 A0A3Q1I1S9 Q803B4 A0A3B5BAG1
PDB
4YXF
E-value=8.472e-14,
Score=186
Ontologies
PATHWAY
GO
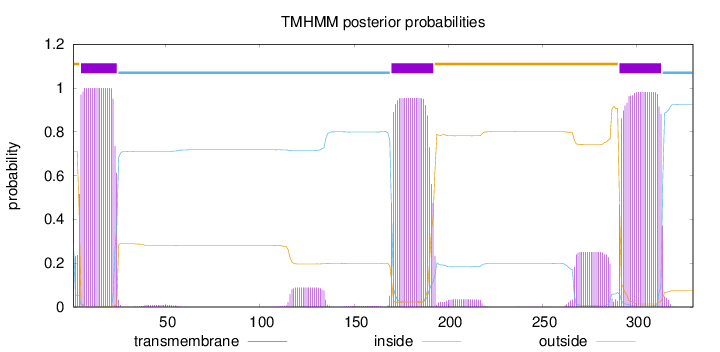
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
330
Number of predicted TMHs:
3
Exp number of AAs in TMHs:
70.52327
Exp number, first 60 AAs:
20.44178
Total prob of N-in:
0.28939
POSSIBLE N-term signal
sequence
outside
1 - 4
TMhelix
5 - 24
inside
25 - 169
TMhelix
170 - 192
outside
193 - 290
TMhelix
291 - 313
inside
314 - 330
Population Genetic Test Statistics
Pi
226.425879
Theta
202.228448
Tajima's D
0.394449
CLR
0.099968
CSRT
0.482725863706815
Interpretation
Possibly Positive selection
