Pre Gene Modal
BGIBMGA011521
Annotation
asparagine_synthetase_[Bombyx_mori]
Full name
Asparagine synthetase [glutamine-hydrolyzing]
Alternative Name
Glutamine-dependent asparagine synthetase
Location in the cell
Cytoplasmic Reliability : 1.86 Mitochondrial Reliability : 1.224
Sequence
CDS
ATGTGCGGTATTTGGGCAACATTCGGTGTAACTGGTGGCCTCACTCCTACCTGCGTAAAATGTTTTTCAAACATTCGCCACCGAGGTCCGGACGCGTGGCGAATTGAGCAAGATGCTAGAGAGCCGTTGGCTATTCTCGGATTTCAACGTCTCGCTATCGTGGACGGTCGGCACGGTATGCAACCGATGCGCCTACACTGTTACCCTCGCACTACGTTAATCTGTAATGGTGAGATTTACAACTGTAAACGCCTACAGAAACAGCATGAGTTTCAATATGAAACAAACTGTGATGTCGAAGCCATCATACATTTGTATCAAAGATTCGGCATAGAAGAAGCCGTCACTAACCTTGACGGGGTTTTTTCGTTTTGCCTTGTCGACGGTGAGAAGAACAAAGTTTATATCGCCCGTGATCCTTATGGCGTTAGACCTCTGTTTAGTTTTCATGACCCTGAAAACGATGTTCTCGCTATTTGTTCTGAAGCTAAAGGCTTGCTCGGCTTAAAGCAAAATGGCAGCCCAAACTCGACACTCGGTCAATTCTCTCCGGGTAACTTACAAGTGTGGAGTATACTTGAAAGTGGTAAAGTCAAACTTGATTACACACATCAATATTTCACACCTGGGAAGGCACCCAAATTTGAGCCTTACGTACCGGAAAATAAATTAATCGATATGAACGTCAATGAAAAGATTGCGTATTTATTGGAGGCCGCTTGCCGAAAACGACTTATGTCCGATCGACGAATAGGCTGTTTACTTAGTGGTGGATTAGATTCCTCGTTAATCACCGCCATTGTAGTGAAACTTGCAAAGGAATACAAATTGCCTTACAAAATACAAACTTTCGCAATCGGGATGGGCGATTCTCCAGATCTTGCAGCCGCCAGAACCGTGGCGGATTATTTAGGAACGGAACATCACGAAGTTCAATTCGATGAAAACGATATAAGAAAAGATCTGGATAACGTTATTTACCATTTAGAATCTTATGATATAACAACTATTCGAGCGAGTTTGCCAATGTACCTTTTGTCAAAGTATATTAAAGAAAAAACAGACACAACAGTGGTATTTAGCGGAGAAGGAGCCGATGAACTAGCGCAAGGCTATATCTACTTCAGAGACGCACCGTCTGAAAAGGACGCACATAAAGAGAGCGTACGTTTGCTTTCTGATATTTATCTCTATGATGGACTGCGAGCAGATAGAACGACGAGTGCCTTTAGTTTAGAATTACGAGTACCGTTTTTAGATATCCAATTCACTCATCACTATCTTTCAATACCTCCTAAACTACGACAACCCCAAAACGGCGTTGAGAAACATCTGTTAAGAAGTAGTTTTGCAAAAAGTGGATTATTGCCAGACTGTGTGCTGTGGCGACACAAAGAAGCATTTAGTGATGGTGTGTCTTCAGTTAAAAAATCGTTGTTCTCGGTCATAAAAGAGATCACAACTGAAAGATTACAAGAAGACGATATACAATATCCAGGACTGCAGCCGAAAACGAATGAATCCAAATATTACCGCTACGTATTTGAGAAATCATTTCCAGGACAACATAATTTTACTCCGTACTATTGGATGCCAAAATGGGTTCAGGTTTCAGACCCTTCGGCAAGGTTTATTAAACATTATGCGGCAAAATGA
Protein
MCGIWATFGVTGGLTPTCVKCFSNIRHRGPDAWRIEQDAREPLAILGFQRLAIVDGRHGMQPMRLHCYPRTTLICNGEIYNCKRLQKQHEFQYETNCDVEAIIHLYQRFGIEEAVTNLDGVFSFCLVDGEKNKVYIARDPYGVRPLFSFHDPENDVLAICSEAKGLLGLKQNGSPNSTLGQFSPGNLQVWSILESGKVKLDYTHQYFTPGKAPKFEPYVPENKLIDMNVNEKIAYLLEAACRKRLMSDRRIGCLLSGGLDSSLITAIVVKLAKEYKLPYKIQTFAIGMGDSPDLAAARTVADYLGTEHHEVQFDENDIRKDLDNVIYHLESYDITTIRASLPMYLLSKYIKEKTDTTVVFSGEGADELAQGYIYFRDAPSEKDAHKESVRLLSDIYLYDGLRADRTTSAFSLELRVPFLDIQFTHHYLSIPPKLRQPQNGVEKHLLRSSFAKSGLLPDCVLWRHKEAFSDGVSSVKKSLFSVIKEITTERLQEDDIQYPGLQPKTNESKYYRYVFEKSFPGQHNFTPYYWMPKWVQVSDPSARFIKHYAAK
Summary
Catalytic Activity
ATP + H2O + L-aspartate + L-glutamine = AMP + diphosphate + H(+) + L-asparagine + L-glutamate
Cofactor
Mn(2+)
Miscellaneous
The temperature-sensitive mutation affecting G1 progression results from a single amino acid substitution.
Similarity
Belongs to the glycosyltransferase 2 family. GalNAc-T subfamily.
Keywords
Acetylation
Amino-acid biosynthesis
Asparagine biosynthesis
ATP-binding
Glutamine amidotransferase
Ligase
Nucleotide-binding
Phosphoprotein
Complete proteome
Reference proteome
Feature
chain Asparagine synthetase [glutamine-hydrolyzing]
Uniprot
H9JPR6
Q2L9S5
A0A194RR70
A0A194PSC8
S4PMX8
A0A3G1T167
+ More
A0A0L7KX40 A0A2H1V6T3 A0A212FFL1 A0A151X8W9 F4X3F1 E9IBX6 A0A195CJ11 A0A158NKV4 A0A195BLW4 A0A067RI79 A0A195DG80 A0A0J7L123 A0A026VVC4 A0A2J7QC70 A0A2P8XMC8 E2BXP2 E2AJW0 A0A2A3EAF7 A0A1B6MEB6 K7ILZ5 A0A232F6W7 J3JXG4 A0A0A9Y0E6 A0A195FCU3 N6U6K6 A0A1W4XK19 A0A1Y1M6J0 A0A139WI23 A0A224XMM5 A0A0V0G3T6 A0A0P4VKA9 E0VXU0 T1IAZ5 A0A2H8TQ21 A0A2R7W4X3 A0A0L7RFS1 A0A2S2N865 A0A0T6AV08 J9JMX7 A0A0C9R706 T1JD29 A0A1S3INM5 V4ASQ3 A0A2T7PBS1 A0A3S0ZWP2 A0A2D4LJS2 A0A146M7S5 W5MYW5 A0A1W7RJ78 J3SE12 T1E786 A0A098M1N3 A0A0B8RZN1 U3EUA1 R7TGJ1 A0A3S1A575 A0A1W0WEP1 A0A210PVE5 K1PWQ4 M7B7L1 K7F8X7 A0A3Q3GVD8 G3I3J1 P19891 A0A3L7IKG1 A0A1S3ICI2 A0A0B6Z4D5 A0A3N0Y0M4 H0VFI5 I3KUD9 A0A2D0S270 P49088 W5U727 A0A286XA60 A0A1A8IH31 B5X2U4 A0A340WVH0 A0A1U7Q2I5 P17714 A0A2U4AR81 A3KQK2 A0A383YPQ6 Q7ZTW9 A0A2I4CL75 V9KLU5 H2LY62 A0A3P9MBJ8 M4AR02 Q61024 A0A1A8KC68 A0A1A8BTC2 A0A1A8VGN5 A0A091T1K5 A0A341D7L1
A0A0L7KX40 A0A2H1V6T3 A0A212FFL1 A0A151X8W9 F4X3F1 E9IBX6 A0A195CJ11 A0A158NKV4 A0A195BLW4 A0A067RI79 A0A195DG80 A0A0J7L123 A0A026VVC4 A0A2J7QC70 A0A2P8XMC8 E2BXP2 E2AJW0 A0A2A3EAF7 A0A1B6MEB6 K7ILZ5 A0A232F6W7 J3JXG4 A0A0A9Y0E6 A0A195FCU3 N6U6K6 A0A1W4XK19 A0A1Y1M6J0 A0A139WI23 A0A224XMM5 A0A0V0G3T6 A0A0P4VKA9 E0VXU0 T1IAZ5 A0A2H8TQ21 A0A2R7W4X3 A0A0L7RFS1 A0A2S2N865 A0A0T6AV08 J9JMX7 A0A0C9R706 T1JD29 A0A1S3INM5 V4ASQ3 A0A2T7PBS1 A0A3S0ZWP2 A0A2D4LJS2 A0A146M7S5 W5MYW5 A0A1W7RJ78 J3SE12 T1E786 A0A098M1N3 A0A0B8RZN1 U3EUA1 R7TGJ1 A0A3S1A575 A0A1W0WEP1 A0A210PVE5 K1PWQ4 M7B7L1 K7F8X7 A0A3Q3GVD8 G3I3J1 P19891 A0A3L7IKG1 A0A1S3ICI2 A0A0B6Z4D5 A0A3N0Y0M4 H0VFI5 I3KUD9 A0A2D0S270 P49088 W5U727 A0A286XA60 A0A1A8IH31 B5X2U4 A0A340WVH0 A0A1U7Q2I5 P17714 A0A2U4AR81 A3KQK2 A0A383YPQ6 Q7ZTW9 A0A2I4CL75 V9KLU5 H2LY62 A0A3P9MBJ8 M4AR02 Q61024 A0A1A8KC68 A0A1A8BTC2 A0A1A8VGN5 A0A091T1K5 A0A341D7L1
EC Number
6.3.5.4
Pubmed
19121390
26354079
23622113
26227816
22118469
21719571
+ More
21282665 21347285 24845553 24508170 30249741 29403074 20798317 20075255 28648823 22516182 25401762 23537049 28004739 18362917 19820115 27129103 20566863 26383154 23254933 26823975 26358130 23025625 23758969 25727380 25476704 23915248 28812685 22992520 23624526 17381049 21804562 23929341 2477309 29704459 21993624 25186727 7818476 15489334 23127152 20433749 1972978 23594743 24402279 17554307 23542700 16141072 21183079 24139043
21282665 21347285 24845553 24508170 30249741 29403074 20798317 20075255 28648823 22516182 25401762 23537049 28004739 18362917 19820115 27129103 20566863 26383154 23254933 26823975 26358130 23025625 23758969 25727380 25476704 23915248 28812685 22992520 23624526 17381049 21804562 23929341 2477309 29704459 21993624 25186727 7818476 15489334 23127152 20433749 1972978 23594743 24402279 17554307 23542700 16141072 21183079 24139043
EMBL
BABH01015193
DQ343760
ABC69171.1
KQ459833
KPJ19805.1
KQ459601
+ More
KPI94030.1 GAIX01003305 JAA89255.1 MG846873 AXY94725.1 JTDY01004893 KOB67626.1 ODYU01000963 SOQ36509.1 AGBW02008806 OWR52518.1 KQ982409 KYQ56826.1 GL888613 EGI58979.1 GL762137 EFZ22002.1 KQ977649 KYN00731.1 ADTU01018949 KQ976439 KYM86719.1 KK852665 KDR18971.1 KQ980886 KYN11847.1 LBMM01001428 KMQ96336.1 KK107796 QOIP01000013 EZA47738.1 RLU15169.1 NEVH01016291 PNF26186.1 PYGN01001729 PSN33161.1 GL451281 EFN79560.1 GL440100 EFN66276.1 KZ288311 PBC28454.1 GEBQ01005699 JAT34278.1 NNAY01000772 OXU26596.1 BT127933 AEE62895.1 GBHO01020589 JAG23015.1 KQ981693 KYN37864.1 APGK01040817 KB740984 KB631899 ENN76286.1 ERL86933.1 GEZM01042355 GEZM01042354 GEZM01042353 GEZM01042352 JAV79915.1 KQ971343 KYB27475.1 GFTR01007177 JAW09249.1 GECL01003425 JAP02699.1 GDKW01001849 JAI54746.1 DS235840 EEB18196.1 ACPB03000844 GFXV01003543 MBW15348.1 KK854219 PTY13265.1 KQ414601 KOC69684.1 GGMR01000659 MBY13278.1 LJIG01022749 KRT78921.1 ABLF02023962 ABLF02023963 GBYB01012114 JAG81881.1 JH432094 KB201370 ESO96771.1 PZQS01000005 PVD30867.1 RQTK01000080 RUS88523.1 IACM01031018 LAB21307.1 GDHC01002798 JAQ15831.1 AHAT01004624 GDAY02000209 JAV51204.1 JU173940 GBEX01000289 AFJ49466.1 JAI14271.1 GAAZ01000221 GBKC01000298 GBKD01000194 JAA97722.1 JAG45772.1 JAG47424.1 GBSI01000280 JAC96215.1 GBSH01000351 JAG68673.1 GAEP01000173 GBEW01000100 JAB54648.1 JAI10265.1 AMQN01013089 KB309954 ELT92908.1 RQTK01000021 RUS90971.1 MTYJ01000119 OQV13659.1 NEDP02005461 OWF40463.1 JH815782 EKC26133.1 KB534798 EMP33931.1 AGCU01029125 AGCU01029126 AGCU01029127 AGCU01029128 AGCU01029129 AGCU01029130 JH001188 KE663634 EGW08354.1 ERE91184.1 M27838 X12950 RAZU01000039 RLQ78470.1 HACG01016377 CEK63242.1 RJVU01057109 ROK35680.1 AAKN02016952 AAKN02016953 AERX01064707 AERX01064708 AERX01064709 AERX01064710 U07202 U07201 BC081719 JT406840 AHH37746.1 HAED01010057 SBQ96269.1 BT045363 BT059666 ACI33625.1 ACN11379.1 X52130 BX842688 BC052127 AAH52127.1 JW866458 AFO98975.1 U38940 AK076207 AK167524 BC005552 HAEE01009209 SBR29259.1 HADZ01005829 HAEA01009993 SBP69770.1 HADY01014562 HAEJ01018584 SBS59041.1 KK940410 KFQ50258.1
KPI94030.1 GAIX01003305 JAA89255.1 MG846873 AXY94725.1 JTDY01004893 KOB67626.1 ODYU01000963 SOQ36509.1 AGBW02008806 OWR52518.1 KQ982409 KYQ56826.1 GL888613 EGI58979.1 GL762137 EFZ22002.1 KQ977649 KYN00731.1 ADTU01018949 KQ976439 KYM86719.1 KK852665 KDR18971.1 KQ980886 KYN11847.1 LBMM01001428 KMQ96336.1 KK107796 QOIP01000013 EZA47738.1 RLU15169.1 NEVH01016291 PNF26186.1 PYGN01001729 PSN33161.1 GL451281 EFN79560.1 GL440100 EFN66276.1 KZ288311 PBC28454.1 GEBQ01005699 JAT34278.1 NNAY01000772 OXU26596.1 BT127933 AEE62895.1 GBHO01020589 JAG23015.1 KQ981693 KYN37864.1 APGK01040817 KB740984 KB631899 ENN76286.1 ERL86933.1 GEZM01042355 GEZM01042354 GEZM01042353 GEZM01042352 JAV79915.1 KQ971343 KYB27475.1 GFTR01007177 JAW09249.1 GECL01003425 JAP02699.1 GDKW01001849 JAI54746.1 DS235840 EEB18196.1 ACPB03000844 GFXV01003543 MBW15348.1 KK854219 PTY13265.1 KQ414601 KOC69684.1 GGMR01000659 MBY13278.1 LJIG01022749 KRT78921.1 ABLF02023962 ABLF02023963 GBYB01012114 JAG81881.1 JH432094 KB201370 ESO96771.1 PZQS01000005 PVD30867.1 RQTK01000080 RUS88523.1 IACM01031018 LAB21307.1 GDHC01002798 JAQ15831.1 AHAT01004624 GDAY02000209 JAV51204.1 JU173940 GBEX01000289 AFJ49466.1 JAI14271.1 GAAZ01000221 GBKC01000298 GBKD01000194 JAA97722.1 JAG45772.1 JAG47424.1 GBSI01000280 JAC96215.1 GBSH01000351 JAG68673.1 GAEP01000173 GBEW01000100 JAB54648.1 JAI10265.1 AMQN01013089 KB309954 ELT92908.1 RQTK01000021 RUS90971.1 MTYJ01000119 OQV13659.1 NEDP02005461 OWF40463.1 JH815782 EKC26133.1 KB534798 EMP33931.1 AGCU01029125 AGCU01029126 AGCU01029127 AGCU01029128 AGCU01029129 AGCU01029130 JH001188 KE663634 EGW08354.1 ERE91184.1 M27838 X12950 RAZU01000039 RLQ78470.1 HACG01016377 CEK63242.1 RJVU01057109 ROK35680.1 AAKN02016952 AAKN02016953 AERX01064707 AERX01064708 AERX01064709 AERX01064710 U07202 U07201 BC081719 JT406840 AHH37746.1 HAED01010057 SBQ96269.1 BT045363 BT059666 ACI33625.1 ACN11379.1 X52130 BX842688 BC052127 AAH52127.1 JW866458 AFO98975.1 U38940 AK076207 AK167524 BC005552 HAEE01009209 SBR29259.1 HADZ01005829 HAEA01009993 SBP69770.1 HADY01014562 HAEJ01018584 SBS59041.1 KK940410 KFQ50258.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000037510
UP000007151
UP000075809
+ More
UP000007755 UP000078542 UP000005205 UP000078540 UP000027135 UP000078492 UP000036403 UP000053097 UP000279307 UP000235965 UP000245037 UP000008237 UP000000311 UP000242457 UP000002358 UP000215335 UP000078541 UP000019118 UP000030742 UP000192223 UP000007266 UP000009046 UP000015103 UP000053825 UP000007819 UP000085678 UP000030746 UP000245119 UP000271974 UP000018468 UP000014760 UP000242188 UP000005408 UP000031443 UP000007267 UP000261660 UP000001075 UP000030759 UP000273346 UP000005447 UP000005207 UP000221080 UP000002494 UP000087266 UP000265300 UP000189706 UP000245320 UP000000437 UP000261681 UP000192220 UP000001038 UP000265180 UP000002852 UP000000589 UP000252040
UP000007755 UP000078542 UP000005205 UP000078540 UP000027135 UP000078492 UP000036403 UP000053097 UP000279307 UP000235965 UP000245037 UP000008237 UP000000311 UP000242457 UP000002358 UP000215335 UP000078541 UP000019118 UP000030742 UP000192223 UP000007266 UP000009046 UP000015103 UP000053825 UP000007819 UP000085678 UP000030746 UP000245119 UP000271974 UP000018468 UP000014760 UP000242188 UP000005408 UP000031443 UP000007267 UP000261660 UP000001075 UP000030759 UP000273346 UP000005447 UP000005207 UP000221080 UP000002494 UP000087266 UP000265300 UP000189706 UP000245320 UP000000437 UP000261681 UP000192220 UP000001038 UP000265180 UP000002852 UP000000589 UP000252040
Pfam
Interpro
IPR017932
GATase_2_dom
+ More
IPR029055 Ntn_hydrolases_N
IPR001962 Asn_synthase
IPR014729 Rossmann-like_a/b/a_fold
IPR006426 Asn_synth_AEB
IPR033738 AsnB_N
IPR012337 RNaseH-like_sf
IPR013520 Exonuclease_RNaseT/DNA_pol3
IPR022894 Oligoribonuclease
IPR036397 RNaseH_sf
IPR001173 Glyco_trans_2-like
IPR029044 Nucleotide-diphossugar_trans
IPR035992 Ricin_B-like_lectins
IPR000772 Ricin_B_lectin
IPR028045 DUF4539
IPR029055 Ntn_hydrolases_N
IPR001962 Asn_synthase
IPR014729 Rossmann-like_a/b/a_fold
IPR006426 Asn_synth_AEB
IPR033738 AsnB_N
IPR012337 RNaseH-like_sf
IPR013520 Exonuclease_RNaseT/DNA_pol3
IPR022894 Oligoribonuclease
IPR036397 RNaseH_sf
IPR001173 Glyco_trans_2-like
IPR029044 Nucleotide-diphossugar_trans
IPR035992 Ricin_B-like_lectins
IPR000772 Ricin_B_lectin
IPR028045 DUF4539
Gene 3D
ProteinModelPortal
H9JPR6
Q2L9S5
A0A194RR70
A0A194PSC8
S4PMX8
A0A3G1T167
+ More
A0A0L7KX40 A0A2H1V6T3 A0A212FFL1 A0A151X8W9 F4X3F1 E9IBX6 A0A195CJ11 A0A158NKV4 A0A195BLW4 A0A067RI79 A0A195DG80 A0A0J7L123 A0A026VVC4 A0A2J7QC70 A0A2P8XMC8 E2BXP2 E2AJW0 A0A2A3EAF7 A0A1B6MEB6 K7ILZ5 A0A232F6W7 J3JXG4 A0A0A9Y0E6 A0A195FCU3 N6U6K6 A0A1W4XK19 A0A1Y1M6J0 A0A139WI23 A0A224XMM5 A0A0V0G3T6 A0A0P4VKA9 E0VXU0 T1IAZ5 A0A2H8TQ21 A0A2R7W4X3 A0A0L7RFS1 A0A2S2N865 A0A0T6AV08 J9JMX7 A0A0C9R706 T1JD29 A0A1S3INM5 V4ASQ3 A0A2T7PBS1 A0A3S0ZWP2 A0A2D4LJS2 A0A146M7S5 W5MYW5 A0A1W7RJ78 J3SE12 T1E786 A0A098M1N3 A0A0B8RZN1 U3EUA1 R7TGJ1 A0A3S1A575 A0A1W0WEP1 A0A210PVE5 K1PWQ4 M7B7L1 K7F8X7 A0A3Q3GVD8 G3I3J1 P19891 A0A3L7IKG1 A0A1S3ICI2 A0A0B6Z4D5 A0A3N0Y0M4 H0VFI5 I3KUD9 A0A2D0S270 P49088 W5U727 A0A286XA60 A0A1A8IH31 B5X2U4 A0A340WVH0 A0A1U7Q2I5 P17714 A0A2U4AR81 A3KQK2 A0A383YPQ6 Q7ZTW9 A0A2I4CL75 V9KLU5 H2LY62 A0A3P9MBJ8 M4AR02 Q61024 A0A1A8KC68 A0A1A8BTC2 A0A1A8VGN5 A0A091T1K5 A0A341D7L1
A0A0L7KX40 A0A2H1V6T3 A0A212FFL1 A0A151X8W9 F4X3F1 E9IBX6 A0A195CJ11 A0A158NKV4 A0A195BLW4 A0A067RI79 A0A195DG80 A0A0J7L123 A0A026VVC4 A0A2J7QC70 A0A2P8XMC8 E2BXP2 E2AJW0 A0A2A3EAF7 A0A1B6MEB6 K7ILZ5 A0A232F6W7 J3JXG4 A0A0A9Y0E6 A0A195FCU3 N6U6K6 A0A1W4XK19 A0A1Y1M6J0 A0A139WI23 A0A224XMM5 A0A0V0G3T6 A0A0P4VKA9 E0VXU0 T1IAZ5 A0A2H8TQ21 A0A2R7W4X3 A0A0L7RFS1 A0A2S2N865 A0A0T6AV08 J9JMX7 A0A0C9R706 T1JD29 A0A1S3INM5 V4ASQ3 A0A2T7PBS1 A0A3S0ZWP2 A0A2D4LJS2 A0A146M7S5 W5MYW5 A0A1W7RJ78 J3SE12 T1E786 A0A098M1N3 A0A0B8RZN1 U3EUA1 R7TGJ1 A0A3S1A575 A0A1W0WEP1 A0A210PVE5 K1PWQ4 M7B7L1 K7F8X7 A0A3Q3GVD8 G3I3J1 P19891 A0A3L7IKG1 A0A1S3ICI2 A0A0B6Z4D5 A0A3N0Y0M4 H0VFI5 I3KUD9 A0A2D0S270 P49088 W5U727 A0A286XA60 A0A1A8IH31 B5X2U4 A0A340WVH0 A0A1U7Q2I5 P17714 A0A2U4AR81 A3KQK2 A0A383YPQ6 Q7ZTW9 A0A2I4CL75 V9KLU5 H2LY62 A0A3P9MBJ8 M4AR02 Q61024 A0A1A8KC68 A0A1A8BTC2 A0A1A8VGN5 A0A091T1K5 A0A341D7L1
PDB
1CT9
E-value=4.15162e-73,
Score=700
Ontologies
PATHWAY
GO
GO:0004066
GO:0006529
GO:0005524
GO:0006541
GO:0000175
GO:0003676
GO:0005794
GO:0016021
GO:0006486
GO:0030246
GO:0016757
GO:0005829
GO:0045931
GO:0043066
GO:0042149
GO:0070981
GO:0032354
GO:0031427
GO:0009416
GO:0001889
GO:0031667
GO:0009636
GO:0043200
GO:0009612
GO:0006520
GO:0048037
GO:0032870
GO:0042803
GO:0031625
GO:0005515
GO:0000287
GO:0016620
GO:0016787
GO:0016491
GO:0015930
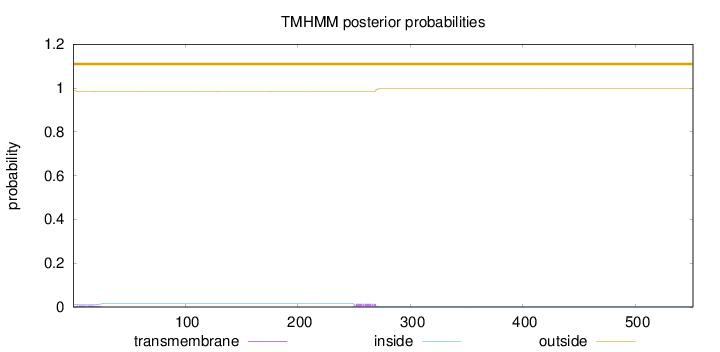
Topology
Length:
551
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.419500000000001
Exp number, first 60 AAs:
0.12754
Total prob of N-in:
0.01216
outside
1 - 551
Population Genetic Test Statistics
Pi
155.579539
Theta
186.764735
Tajima's D
-0.637383
CLR
141.874101
CSRT
0.213239338033098
Interpretation
Possibly Positive selection
