Gene
KWMTBOMO13743
Pre Gene Modal
BGIBMGA014468
Annotation
non-LTR_retrotransposon_CATS_[Danaus_plexippus]
Location in the cell
Mitochondrial Reliability : 1.666
Sequence
CDS
ATGGCAATCCATCTACCATCTGGATGCCACATGCAGGCCTACGCAGACGATGTGGTACTCGTGGTCACCGCCGAAAATGTCAATCGCCTAGAGGAGCTCACCGACACGGTTCTCCAACAGATAGTGAATTGGGGAAAAAGCGTCAAGCTCGAATTCGGGGCCTTCAAAACACAACTGATCGCCTTCACCCCCAAGGCCAGAACTCTTAGGGTGGAGATGGACGGTCAAATCTTAAAAACCTGTAAGGAGATCAAGCTCCTCGGAATAATTTTGGACGAAAAGCTGCTCTTTGGCCGCCATGTACAGTACGTCCTGGGAAAAGCCTCGCGAATATTCAACAAACTGTGCTTGTACTCCCGACCAACCTGGGGTGCTCATCCCGAGAACGTGCGCACCATCTATCTCCGAGTCATCGAGCCAATCGTCACCTACGCTGCCGGGGTGTGGGGACACATAGTAAACAAGCGCTACATCAAAAAATCTTTGATGAGCATGCAGCGTGGTTTTGCCCTAAAAGCCATCAGAGGATTCCGCACTATTTCAACCTCCGCTGCTCTGGCTCTCGCACAATTCACCCCTCTAGACCTAAAAATCCGAGAAGTACACCAAATCGAAAAAGTCAGGCTCACTGGGAAAACTGAATTCCTTCCCAATGACATTTCTTATGAGAAACCTGTCCCCGTCAAGGACCTCCTACATCCCGCTAGAAGGCCGACTATCAAACCCATACCCTCCTCTGAACACCGAATTAACTCCGAAATGACGGACAGTACTCTGGTCACTCGCGTATTCACTGACGGAAGTAAGCAGGACGATGGATCAGTTGGAGCCGCATTCGTGCTTTACAGACCCGGCGACACTGAAACATCCGTAACAAAAAAGTACAAATTACACAGTAGCTGCACAGTCTTCCAGGCAGAGCTGTACGCCATCTGGAAAGCCTGTCAGTGGATTGTAAGCGAAAATCATCCCCACACATACATCTTCACCGATTCTCTCTCATCAATATACGCGATAAATAACAGAAGCAACACACACCCCATCATAGTGAACATACACAAACTCATACACGCTAACCACACTACACACAAAATTTACATAGATTGGGTAAAGGGACACTCGGGTGTCATAGGAAACGAGGAAGCCGATGCGGCTGCCAACAGCGGCGCACGCATGCACAAGGCCCCCGACTACTCGCAGTTCTCAATATCCTACGTGAAACACAAGATTCGCACAGAACACCACGCCATCTGGCAAGCCAGATACGAAAGCTCCCCGCAGGGCTCACACACAAAAACACTACTACCAAAACTGAACGACATCAAAGAACTTAACAAATGCACACAAACAAACTTTCAGCATACACAAATACTCACAGGCCATGGATATCACAAGACATACTTACATCGCTTCAAAATAGCACCAGATGACACCTGCCCCTGCGACGGCACCTCCAGCCAAACTGTAGAACACCTCCTTAAAAACTGCCAAGCGTTCGCTGCGAAAAGGCACAGTCACGAAGAAACGTGCCACGCAGTACAGGTGCCACCATATAATTTACCTGAAATGTTAAAAAAGAAAGCTGCAATCAACTCTTTCACCTCGTTTTGCCAAACAATAATCAACAACATCAAGAAAATAAATGAAAACAATTAG
Protein
MAIHLPSGCHMQAYADDVVLVVTAENVNRLEELTDTVLQQIVNWGKSVKLEFGAFKTQLIAFTPKARTLRVEMDGQILKTCKEIKLLGIILDEKLLFGRHVQYVLGKASRIFNKLCLYSRPTWGAHPENVRTIYLRVIEPIVTYAAGVWGHIVNKRYIKKSLMSMQRGFALKAIRGFRTISTSAALALAQFTPLDLKIREVHQIEKVRLTGKTEFLPNDISYEKPVPVKDLLHPARRPTIKPIPSSEHRINSEMTDSTLVTRVFTDGSKQDDGSVGAAFVLYRPGDTETSVTKKYKLHSSCTVFQAELYAIWKACQWIVSENHPHTYIFTDSLSSIYAINNRSNTHPIIVNIHKLIHANHTTHKIYIDWVKGHSGVIGNEEADAAANSGARMHKAPDYSQFSISYVKHKIRTEHHAIWQARYESSPQGSHTKTLLPKLNDIKELNKCTQTNFQHTQILTGHGYHKTYLHRFKIAPDDTCPCDGTSSQTVEHLLKNCQAFAAKRHSHEETCHAVQVPPYNLPEMLKKKAAINSFTSFCQTIINNIKKINENN
Summary
Uniprot
A0A3S2NBZ7
Q9BLI5
Q3V5Y4
Q93139
A0A3S3P0A9
A0A2A4JF48
+ More
A0A1W7R9Y4 A0A2W1BN41 A0A2L2YM05 A0A224X5M4 A0A087TAD8 A0A147BK23 A0A1Y3BBS5 A0A0K1IK29 A0A087TJZ1 A0A0K1IK14 A0A087UHT1 A0A087TI44 A0A0K8TSF0 A0A087USZ4 A0A131XRZ7 O44317 L7MBK1 A0A1B6G0A1 A0A142LX31 A0A023EX80 O44315 A0A087TUG6 A0A131XRY9 A0A131Y6M1 A0A131XWC0 O46185 U5EVA2 U5EV73 A0A2E0U2U9 U5EW32 A0A224X5X2 A0A1W4XBW8 A0A224X6B3 A0A142LX39 A0A0G2LA86 U5EG91 U5ERW4 A0A2R5L945 A0A023EWA3 A0A142LX33 K7JA12 A0A0K8TR49 A0A0K8TRR6 A0A3S2KYU3 K7JB14 A0A0J7KB01 A0A2P2I3J0 A0A1B6ETM1 O18558 A0A0K8TR39 A0A142LX24 U5EV16 U5ERR6 C5KEL8 U5EFQ1 X1WW00 J9MAR8 A0A2S2QV27 A0A0A2KN74 A0A1M2W257 A0A224X696 A0A074RHS1 X8JC12 V9H1I9 A0A226DPQ5 A0A2S2Q1Q6 M5CE34 A0A0G4PXY0 U5EPW8 A0A1S4EP80 A0A1Q5UHD9
A0A1W7R9Y4 A0A2W1BN41 A0A2L2YM05 A0A224X5M4 A0A087TAD8 A0A147BK23 A0A1Y3BBS5 A0A0K1IK29 A0A087TJZ1 A0A0K1IK14 A0A087UHT1 A0A087TI44 A0A0K8TSF0 A0A087USZ4 A0A131XRZ7 O44317 L7MBK1 A0A1B6G0A1 A0A142LX31 A0A023EX80 O44315 A0A087TUG6 A0A131XRY9 A0A131Y6M1 A0A131XWC0 O46185 U5EVA2 U5EV73 A0A2E0U2U9 U5EW32 A0A224X5X2 A0A1W4XBW8 A0A224X6B3 A0A142LX39 A0A0G2LA86 U5EG91 U5ERW4 A0A2R5L945 A0A023EWA3 A0A142LX33 K7JA12 A0A0K8TR49 A0A0K8TRR6 A0A3S2KYU3 K7JB14 A0A0J7KB01 A0A2P2I3J0 A0A1B6ETM1 O18558 A0A0K8TR39 A0A142LX24 U5EV16 U5ERR6 C5KEL8 U5EFQ1 X1WW00 J9MAR8 A0A2S2QV27 A0A0A2KN74 A0A1M2W257 A0A224X696 A0A074RHS1 X8JC12 V9H1I9 A0A226DPQ5 A0A2S2Q1Q6 M5CE34 A0A0G4PXY0 U5EPW8 A0A1S4EP80 A0A1Q5UHD9
Pubmed
EMBL
RSAL01000249
RVE43511.1
AB046668
BAB21511.1
AB211537
BAE44464.1
+ More
D38414 BAA07467.1 NCKU01007943 RWS02258.1 NWSH01001820 PCG70022.1 GFAH01000444 JAV47945.1 KZ149985 PZC75691.1 IAAA01038678 LAA08500.1 GFTR01008639 JAW07787.1 KK114278 KFM62077.1 GEGO01004274 JAR91130.1 MUJZ01028333 OTF78340.1 KP771711 AKU04648.1 KK115566 KFM65430.1 KP771712 AKU04650.1 KK119855 KFM76920.1 KK115320 KFM64783.1 GDAI01000743 JAI16860.1 KK121445 KFM80483.1 GEFM01006379 JAP69417.1 AF015813 AAB94039.1 GACK01003634 JAA61400.1 GECZ01013919 JAS55850.1 KU543676 AMS38357.1 GBBI01004920 JAC13792.1 AF015489 AAB94030.1 KK116791 KFM68755.1 GEFM01006389 JAP69407.1 GEFM01002205 JAP73591.1 GEFM01005915 JAP69881.1 U73803 AAB92394.1 GANO01003496 JAB56375.1 GANO01003537 JAB56334.1 PABS01000003 MAT33547.1 GANO01003141 JAB56730.1 GFTR01008541 JAW07885.1 GFTR01008411 JAW08015.1 KU543680 AMS38365.1 CR848788 GANO01003539 JAB56332.1 GANO01003538 JAB56333.1 GGLE01001887 MBY06013.1 GAPW01000103 JAC13495.1 KU543677 AMS38359.1 AAZX01016607 AAZX01023407 GDAI01000751 JAI16852.1 GDAI01000752 JAI16851.1 RSAL01000438 RVE41707.1 AAZX01002319 LBMM01010475 KMQ87436.1 IACF01002963 LAB68595.1 GECZ01028519 JAS41250.1 U87543 AAB65093.1 GDAI01000756 JAI16847.1 KU543672 AMS38350.1 GANO01003611 JAB56260.1 GANO01003612 JAB56259.1 GG672330 EER17156.1 GANO01003746 JAB56125.1 ABLF02060681 ABLF02064366 ABLF02030052 GGMS01012395 MBY81598.1 JQGA01001155 KGO69287.1 MNAD01000345 OJT13951.1 GFTR01008431 JAW07995.1 AZST01001167 KEP46314.1 JATN01000319 EUC61565.1 X59239 CAA41925.1 LNIX01000015 OXA46641.1 GGMS01001879 MBY71082.1 CAOJ01012627 LN679168 CCO34112.1 CEL62418.1 HG793200 CRL30999.1 GANO01003536 JAB56335.1 MNBE01000390 MNBE01000261 OKP10147.1 OKP11873.1
D38414 BAA07467.1 NCKU01007943 RWS02258.1 NWSH01001820 PCG70022.1 GFAH01000444 JAV47945.1 KZ149985 PZC75691.1 IAAA01038678 LAA08500.1 GFTR01008639 JAW07787.1 KK114278 KFM62077.1 GEGO01004274 JAR91130.1 MUJZ01028333 OTF78340.1 KP771711 AKU04648.1 KK115566 KFM65430.1 KP771712 AKU04650.1 KK119855 KFM76920.1 KK115320 KFM64783.1 GDAI01000743 JAI16860.1 KK121445 KFM80483.1 GEFM01006379 JAP69417.1 AF015813 AAB94039.1 GACK01003634 JAA61400.1 GECZ01013919 JAS55850.1 KU543676 AMS38357.1 GBBI01004920 JAC13792.1 AF015489 AAB94030.1 KK116791 KFM68755.1 GEFM01006389 JAP69407.1 GEFM01002205 JAP73591.1 GEFM01005915 JAP69881.1 U73803 AAB92394.1 GANO01003496 JAB56375.1 GANO01003537 JAB56334.1 PABS01000003 MAT33547.1 GANO01003141 JAB56730.1 GFTR01008541 JAW07885.1 GFTR01008411 JAW08015.1 KU543680 AMS38365.1 CR848788 GANO01003539 JAB56332.1 GANO01003538 JAB56333.1 GGLE01001887 MBY06013.1 GAPW01000103 JAC13495.1 KU543677 AMS38359.1 AAZX01016607 AAZX01023407 GDAI01000751 JAI16852.1 GDAI01000752 JAI16851.1 RSAL01000438 RVE41707.1 AAZX01002319 LBMM01010475 KMQ87436.1 IACF01002963 LAB68595.1 GECZ01028519 JAS41250.1 U87543 AAB65093.1 GDAI01000756 JAI16847.1 KU543672 AMS38350.1 GANO01003611 JAB56260.1 GANO01003612 JAB56259.1 GG672330 EER17156.1 GANO01003746 JAB56125.1 ABLF02060681 ABLF02064366 ABLF02030052 GGMS01012395 MBY81598.1 JQGA01001155 KGO69287.1 MNAD01000345 OJT13951.1 GFTR01008431 JAW07995.1 AZST01001167 KEP46314.1 JATN01000319 EUC61565.1 X59239 CAA41925.1 LNIX01000015 OXA46641.1 GGMS01001879 MBY71082.1 CAOJ01012627 LN679168 CCO34112.1 CEL62418.1 HG793200 CRL30999.1 GANO01003536 JAB56335.1 MNBE01000390 MNBE01000261 OKP10147.1 OKP11873.1
Proteomes
Pfam
Interpro
Gene 3D
ProteinModelPortal
A0A3S2NBZ7
Q9BLI5
Q3V5Y4
Q93139
A0A3S3P0A9
A0A2A4JF48
+ More
A0A1W7R9Y4 A0A2W1BN41 A0A2L2YM05 A0A224X5M4 A0A087TAD8 A0A147BK23 A0A1Y3BBS5 A0A0K1IK29 A0A087TJZ1 A0A0K1IK14 A0A087UHT1 A0A087TI44 A0A0K8TSF0 A0A087USZ4 A0A131XRZ7 O44317 L7MBK1 A0A1B6G0A1 A0A142LX31 A0A023EX80 O44315 A0A087TUG6 A0A131XRY9 A0A131Y6M1 A0A131XWC0 O46185 U5EVA2 U5EV73 A0A2E0U2U9 U5EW32 A0A224X5X2 A0A1W4XBW8 A0A224X6B3 A0A142LX39 A0A0G2LA86 U5EG91 U5ERW4 A0A2R5L945 A0A023EWA3 A0A142LX33 K7JA12 A0A0K8TR49 A0A0K8TRR6 A0A3S2KYU3 K7JB14 A0A0J7KB01 A0A2P2I3J0 A0A1B6ETM1 O18558 A0A0K8TR39 A0A142LX24 U5EV16 U5ERR6 C5KEL8 U5EFQ1 X1WW00 J9MAR8 A0A2S2QV27 A0A0A2KN74 A0A1M2W257 A0A224X696 A0A074RHS1 X8JC12 V9H1I9 A0A226DPQ5 A0A2S2Q1Q6 M5CE34 A0A0G4PXY0 U5EPW8 A0A1S4EP80 A0A1Q5UHD9
A0A1W7R9Y4 A0A2W1BN41 A0A2L2YM05 A0A224X5M4 A0A087TAD8 A0A147BK23 A0A1Y3BBS5 A0A0K1IK29 A0A087TJZ1 A0A0K1IK14 A0A087UHT1 A0A087TI44 A0A0K8TSF0 A0A087USZ4 A0A131XRZ7 O44317 L7MBK1 A0A1B6G0A1 A0A142LX31 A0A023EX80 O44315 A0A087TUG6 A0A131XRY9 A0A131Y6M1 A0A131XWC0 O46185 U5EVA2 U5EV73 A0A2E0U2U9 U5EW32 A0A224X5X2 A0A1W4XBW8 A0A224X6B3 A0A142LX39 A0A0G2LA86 U5EG91 U5ERW4 A0A2R5L945 A0A023EWA3 A0A142LX33 K7JA12 A0A0K8TR49 A0A0K8TRR6 A0A3S2KYU3 K7JB14 A0A0J7KB01 A0A2P2I3J0 A0A1B6ETM1 O18558 A0A0K8TR39 A0A142LX24 U5EV16 U5ERR6 C5KEL8 U5EFQ1 X1WW00 J9MAR8 A0A2S2QV27 A0A0A2KN74 A0A1M2W257 A0A224X696 A0A074RHS1 X8JC12 V9H1I9 A0A226DPQ5 A0A2S2Q1Q6 M5CE34 A0A0G4PXY0 U5EPW8 A0A1S4EP80 A0A1Q5UHD9
PDB
2QKK
E-value=5.76573e-05,
Score=112
Ontologies
KEGG
GO
PANTHER
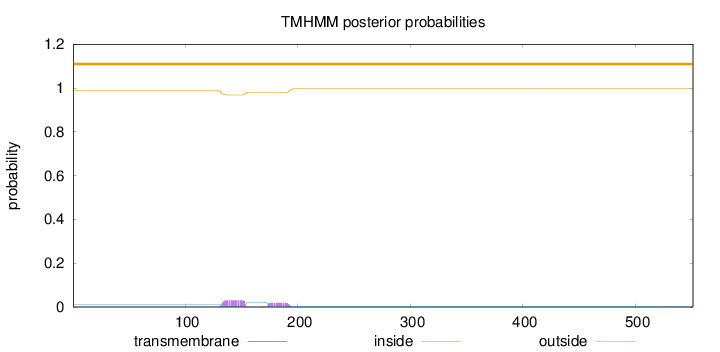
Topology
Length:
551
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.02183
Exp number, first 60 AAs:
0.00029
Total prob of N-in:
0.01258
outside
1 - 551
Population Genetic Test Statistics
Pi
197.512959
Theta
169.476642
Tajima's D
0.346857
CLR
303.603018
CSRT
0.467776611169442
Interpretation
Uncertain
