Gene
KWMTBOMO13732
Annotation
PREDICTED:_protein_TERMINAL_FLOWER_1-like_[Bombyx_mori]
Full name
Protein TERMINAL FLOWER 1
Location in the cell
Extracellular Reliability : 1.628
Sequence
CDS
ATGACTGAAGTGAAAAGTTTACTCGAAGGTTGTGACAGATTGACAGGATTAAACATTACTAGTGTTGGGGGTACAATTGTAAATGATCATAACTGTGATGTTTTACTACCTGCGCAGGTATTTTTGGATGAACCATTATTTCAGTACTTCATGGCTGATTCGAAAAAGTTTTACACGATTATACTGGTGGATCCCGATTCGCCGCCACAAGTAGACGGTGAATTTTATTTGCATATGCTAAAATCGAATATTCCTGGTCTCGCATTGAAAACAAAGGAAAGTAGCAAAACTATTGGAATCGATTACAGAGGATACAAGCCGCCTACTCCATCTCGCGGCATAGATACGCACCGCTACATTACTCTCCTTTATGAGCAAGCAGACGGAAATAATTTTCTACCGACTGTACCGAGCTCACGAAATCGTTTTTCGTTAGCTAAGTGGCTTTTGGGAAAGAACCTTTGCGGCCCGGTCGCTGGAACTCAATTTCGATTGCAATTCTAG
Protein
MTEVKSLLEGCDRLTGLNITSVGGTIVNDHNCDVLLPAQVFLDEPLFQYFMADSKKFYTIILVDPDSPPQVDGEFYLHMLKSNIPGLALKTKESSKTIGIDYRGYKPPTPSRGIDTHRYITLLYEQADGNNFLPTVPSSRNRFSLAKWLLGKNLCGPVAGTQFRLQF
Summary
Description
Controls inflorescence meristem identity and is required for maintenance of an indeterminate inflorescence. Prevents the expression of 'APETALA1' and 'LEAFY'. Also plays a role in the regulation of the time of flowering in the long-day flowering pathway. May form complexes with phosphorylated ligands by interfering with kinases and their effectors (By similarity).
Miscellaneous
Mutagenesis of His-88 to Tyr converts the repressor of flowering "TERMINAL FLOWER 1" into a "FLOWERING LOCUS T"-like activator of flowering.
Similarity
Belongs to the phosphatidylethanolamine-binding protein family.
Keywords
3D-structure
Complete proteome
Cytoplasm
Developmental protein
Differentiation
Flowering
Reference proteome
Feature
chain Protein TERMINAL FLOWER 1
Uniprot
A0A2A4JDP2
A0A2H1X3Q8
A0A212FJL0
A0A194RPL9
A0A3S2P1T2
A0A0L7L8T4
+ More
A0A194PSD3 A0A1B6L590 A0A1B6JCU3 A0A1B6HVJ4 A0A0A9Z6H7 A0A0K8SA16 A0A146KTY3 A0A336LMI4 A0A1B6MGR6 E0VJ75 A0A1B6DB80 A0A182GFY5 A0A1W7R909 A0A023EJ25 A0A336MQQ6 A0A0N5DEJ7 B0WSF7 A0A1Y1N6P1 A0A2H8TK93 B4MBN6 F7EE13 A0A3B4BZI1 A0A3Q1GL66 A0A1B6M2D0 K7IVM0 J9FNI2 U3JZ48 A0A3P7EDL0 K7IVL8 A0A1B6E7K7 A0A1I8EAC3 A0A3B5B678 A0A336LIM0 A0A2S2P882 A0A2K8JLK0 A0A0R3RGP3 Q9VI09 Q1HQH6 A0A232F887 A0A1Q3FSX1 A0A1P6C0D3 A0A3Q3LS05 E3TGE4 R4WFB4 A0A2N9ISZ7 A0A0K8UQS4 A0A3Q0T5Y4 Q16QJ8 A0A0H5SCA5 C0SSN1 A0A218V388 A0A0N4SZJ6 A0A0H5SBL2 G7N5S0 M4CNJ5 A0A2P1BQH0 A0A1B1JEH8 A0A066WGK8 B5G191 B4PVQ0 B5G187 A0A3M0KFR0 J3MBF3 Q298A8 F5HPS3 A0A0S7MD46 A0A147B101 A0A3Q2QA16 A9LLZ5 A0A1A8ITN6 A0A1A8JIX3 A0A1A8NXD4 A0A1A8E8E3 A0A1A7Z8M9 Q8LSG9 A0A384L909 Q547P7 P93003 Q8LSH0 A0A182RZH2 R0HBP3 A0A1B1JEG7 A0A182ILC1 B0LQE2 B4NHB5 F4WAF3 A0A0Q9X5A3 B0LQE7 B4XEW2 B4G3B9 B4XEW0 A0A2P5X051
A0A194PSD3 A0A1B6L590 A0A1B6JCU3 A0A1B6HVJ4 A0A0A9Z6H7 A0A0K8SA16 A0A146KTY3 A0A336LMI4 A0A1B6MGR6 E0VJ75 A0A1B6DB80 A0A182GFY5 A0A1W7R909 A0A023EJ25 A0A336MQQ6 A0A0N5DEJ7 B0WSF7 A0A1Y1N6P1 A0A2H8TK93 B4MBN6 F7EE13 A0A3B4BZI1 A0A3Q1GL66 A0A1B6M2D0 K7IVM0 J9FNI2 U3JZ48 A0A3P7EDL0 K7IVL8 A0A1B6E7K7 A0A1I8EAC3 A0A3B5B678 A0A336LIM0 A0A2S2P882 A0A2K8JLK0 A0A0R3RGP3 Q9VI09 Q1HQH6 A0A232F887 A0A1Q3FSX1 A0A1P6C0D3 A0A3Q3LS05 E3TGE4 R4WFB4 A0A2N9ISZ7 A0A0K8UQS4 A0A3Q0T5Y4 Q16QJ8 A0A0H5SCA5 C0SSN1 A0A218V388 A0A0N4SZJ6 A0A0H5SBL2 G7N5S0 M4CNJ5 A0A2P1BQH0 A0A1B1JEH8 A0A066WGK8 B5G191 B4PVQ0 B5G187 A0A3M0KFR0 J3MBF3 Q298A8 F5HPS3 A0A0S7MD46 A0A147B101 A0A3Q2QA16 A9LLZ5 A0A1A8ITN6 A0A1A8JIX3 A0A1A8NXD4 A0A1A8E8E3 A0A1A7Z8M9 Q8LSG9 A0A384L909 Q547P7 P93003 Q8LSH0 A0A182RZH2 R0HBP3 A0A1B1JEG7 A0A182ILC1 B0LQE2 B4NHB5 F4WAF3 A0A0Q9X5A3 B0LQE7 B4XEW2 B4G3B9 B4XEW0 A0A2P5X051
Pubmed
22118469
26354079
26227816
25401762
26823975
20566863
+ More
26483478 24945155 28884022 28004739 17994087 17495919 20075255 26850696 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17204158 28648823 17885136 20634964 23127152 17510324 22002653 21873998 27385013 17018643 17550304 20360741 23481403 15632085 21431295 17993543 11973317 18273534 27354520 8974397 9108281 9330910 11130714 27862469 15894619 23749190 21719571 18171595 25893780
26483478 24945155 28884022 28004739 17994087 17495919 20075255 26850696 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17204158 28648823 17885136 20634964 23127152 17510324 22002653 21873998 27385013 17018643 17550304 20360741 23481403 15632085 21431295 17993543 11973317 18273534 27354520 8974397 9108281 9330910 11130714 27862469 15894619 23749190 21719571 18171595 25893780
EMBL
NWSH01001920
PCG69674.1
ODYU01013194
SOQ59888.1
AGBW02008253
OWR53937.1
+ More
KQ459833 KPJ19798.1 RSAL01000002 RVE54910.1 JTDY01002289 KOB71734.1 KQ459601 KPI94035.1 GEBQ01021085 JAT18892.1 GECU01010666 JAS97040.1 GECU01029040 JAS78666.1 GBHO01004123 GBHO01004122 JAG39481.1 JAG39482.1 GBRD01015709 GDHC01005350 JAG50117.1 JAQ13279.1 GDHC01019883 JAP98745.1 UFQT01000011 SSX17567.1 GEBQ01004892 JAT35085.1 DS235219 EEB13431.1 GEDC01014354 GEDC01000070 JAS22944.1 JAS37228.1 JXUM01060724 KQ562115 KXJ76655.1 GEHC01000006 JAV47639.1 GAPW01004789 JAC08809.1 UFQS01002176 UFQT01002176 SSX13420.1 SSX32854.1 DS232070 EDS33806.1 GEZM01011439 JAV93521.1 GFXV01002586 MBW14391.1 CH940656 EDW58507.1 GEBQ01009881 JAT30096.1 ADBV01000017 EJW88954.1 AGTO01000187 UYWW01012181 VDM19383.1 GEDC01003388 JAS33910.1 UFQS01004663 UFQT01004663 SSX16539.1 SSX35743.1 GGMR01013040 MBY25659.1 MF683303 ATU82444.1 AE014297 AY075337 AAF54136.1 AAL68202.1 DQ440468 ABF18501.1 NNAY01000752 OXU26688.1 GFDL01004351 JAV30694.1 LN857013 CRZ25717.1 GU589430 ADO29380.1 AB770478 AB839767 BAN13572.1 BAN89466.1 OIVN01006182 SPD27141.1 GDHF01023277 JAI29037.1 CH477746 EAT36672.1 CRZ25716.1 AB435524 BAH37010.1 MUZQ01000064 OWK60191.1 UZAD01000085 VDN82403.1 CRZ25715.1 CM001263 EHH21227.1 MG272468 MG272469 AVI43973.1 KX139001 LR031572 ANS12869.1 VDC78303.1 JMSN01000005 KDN52911.1 DQ215443 ACH45052.1 CM000160 EDW97859.1 ABQF01029470 DQ215439 DQ215440 DQ215445 DQ215446 DQ215447 DQ215448 DQ215449 DQ215451 DQ215452 DQ215453 DQ215454 DQ215455 DQ215459 DQ215461 DQ215462 DQ215463 ACH45048.1 QRBI01000121 RMC05947.1 AP011474 BAX25118.1 CM000070 EAL28047.2 AB562503 BAK23999.1 GBYX01069898 JAO99742.1 GCES01002268 JAR84055.1 EU241912 ABX11023.1 HAED01014261 SBR00706.1 HAEE01000043 SBR20059.1 HAEI01000875 HAEH01009329 SBR73688.1 HADZ01003718 HAEA01014260 SBQ42740.1 HADY01000459 HAEJ01018892 SBP38944.1 AF466812 EU352132 AAM27952.1 ABY79192.1 LUHQ01000005 OAO93664.1 AF466803 AF466804 AF466805 AF466806 AF466808 AF466809 AF466810 AF466811 AF466814 AF466815 AF466816 BT024828 EU352112 EU352113 EU352117 EU352126 EU352127 EU352128 EU352129 EU352130 EU352133 EU352134 KX139000 AAM27944.1 ABD60711.1 ABY79172.1 ANS12868.1 U77674 D86932 D87130 D87519 AB005235 AL162873 CP002688 AF466807 EU352131 AAM27947.1 ABY79191.1 KB870810 EOA22445.1 KX139002 ANS12870.1 EU352115 EU352116 EU352118 EU352119 EU352121 EU352122 EU352123 EU352124 EU352125 ABY79175.1 CH964272 EDW84591.1 GL888048 EGI68836.1 KRG00078.1 EU352120 ABY79180.1 EU026436 EU026443 KM455877 ABW24963.1 ABW24970.1 AIY34696.1 CH479179 EDW24301.1 EU026434 ABW24961.1 KZ665999 PPR96726.1
KQ459833 KPJ19798.1 RSAL01000002 RVE54910.1 JTDY01002289 KOB71734.1 KQ459601 KPI94035.1 GEBQ01021085 JAT18892.1 GECU01010666 JAS97040.1 GECU01029040 JAS78666.1 GBHO01004123 GBHO01004122 JAG39481.1 JAG39482.1 GBRD01015709 GDHC01005350 JAG50117.1 JAQ13279.1 GDHC01019883 JAP98745.1 UFQT01000011 SSX17567.1 GEBQ01004892 JAT35085.1 DS235219 EEB13431.1 GEDC01014354 GEDC01000070 JAS22944.1 JAS37228.1 JXUM01060724 KQ562115 KXJ76655.1 GEHC01000006 JAV47639.1 GAPW01004789 JAC08809.1 UFQS01002176 UFQT01002176 SSX13420.1 SSX32854.1 DS232070 EDS33806.1 GEZM01011439 JAV93521.1 GFXV01002586 MBW14391.1 CH940656 EDW58507.1 GEBQ01009881 JAT30096.1 ADBV01000017 EJW88954.1 AGTO01000187 UYWW01012181 VDM19383.1 GEDC01003388 JAS33910.1 UFQS01004663 UFQT01004663 SSX16539.1 SSX35743.1 GGMR01013040 MBY25659.1 MF683303 ATU82444.1 AE014297 AY075337 AAF54136.1 AAL68202.1 DQ440468 ABF18501.1 NNAY01000752 OXU26688.1 GFDL01004351 JAV30694.1 LN857013 CRZ25717.1 GU589430 ADO29380.1 AB770478 AB839767 BAN13572.1 BAN89466.1 OIVN01006182 SPD27141.1 GDHF01023277 JAI29037.1 CH477746 EAT36672.1 CRZ25716.1 AB435524 BAH37010.1 MUZQ01000064 OWK60191.1 UZAD01000085 VDN82403.1 CRZ25715.1 CM001263 EHH21227.1 MG272468 MG272469 AVI43973.1 KX139001 LR031572 ANS12869.1 VDC78303.1 JMSN01000005 KDN52911.1 DQ215443 ACH45052.1 CM000160 EDW97859.1 ABQF01029470 DQ215439 DQ215440 DQ215445 DQ215446 DQ215447 DQ215448 DQ215449 DQ215451 DQ215452 DQ215453 DQ215454 DQ215455 DQ215459 DQ215461 DQ215462 DQ215463 ACH45048.1 QRBI01000121 RMC05947.1 AP011474 BAX25118.1 CM000070 EAL28047.2 AB562503 BAK23999.1 GBYX01069898 JAO99742.1 GCES01002268 JAR84055.1 EU241912 ABX11023.1 HAED01014261 SBR00706.1 HAEE01000043 SBR20059.1 HAEI01000875 HAEH01009329 SBR73688.1 HADZ01003718 HAEA01014260 SBQ42740.1 HADY01000459 HAEJ01018892 SBP38944.1 AF466812 EU352132 AAM27952.1 ABY79192.1 LUHQ01000005 OAO93664.1 AF466803 AF466804 AF466805 AF466806 AF466808 AF466809 AF466810 AF466811 AF466814 AF466815 AF466816 BT024828 EU352112 EU352113 EU352117 EU352126 EU352127 EU352128 EU352129 EU352130 EU352133 EU352134 KX139000 AAM27944.1 ABD60711.1 ABY79172.1 ANS12868.1 U77674 D86932 D87130 D87519 AB005235 AL162873 CP002688 AF466807 EU352131 AAM27947.1 ABY79191.1 KB870810 EOA22445.1 KX139002 ANS12870.1 EU352115 EU352116 EU352118 EU352119 EU352121 EU352122 EU352123 EU352124 EU352125 ABY79175.1 CH964272 EDW84591.1 GL888048 EGI68836.1 KRG00078.1 EU352120 ABY79180.1 EU026436 EU026443 KM455877 ABW24963.1 ABW24970.1 AIY34696.1 CH479179 EDW24301.1 EU026434 ABW24961.1 KZ665999 PPR96726.1
Proteomes
UP000218220
UP000007151
UP000053240
UP000283053
UP000037510
UP000053268
+ More
UP000009046 UP000069940 UP000249989 UP000046395 UP000002320 UP000008792 UP000002280 UP000261440 UP000257200 UP000002358 UP000004810 UP000016665 UP000270924 UP000093561 UP000261400 UP000050640 UP000000803 UP000215335 UP000006672 UP000261660 UP000221080 UP000261340 UP000008820 UP000197619 UP000038020 UP000278627 UP000011750 UP000027361 UP000002282 UP000007754 UP000269221 UP000006038 UP000001819 UP000265000 UP000078284 UP000006548 UP000075900 UP000029121 UP000075880 UP000007798 UP000007755 UP000189702 UP000008744 UP000239757
UP000009046 UP000069940 UP000249989 UP000046395 UP000002320 UP000008792 UP000002280 UP000261440 UP000257200 UP000002358 UP000004810 UP000016665 UP000270924 UP000093561 UP000261400 UP000050640 UP000000803 UP000215335 UP000006672 UP000261660 UP000221080 UP000261340 UP000008820 UP000197619 UP000038020 UP000278627 UP000011750 UP000027361 UP000002282 UP000007754 UP000269221 UP000006038 UP000001819 UP000265000 UP000078284 UP000006548 UP000075900 UP000029121 UP000075880 UP000007798 UP000007755 UP000189702 UP000008744 UP000239757
PRIDE
Pfam
PF01161 PBP
Interpro
SUPFAM
SSF49777
SSF49777
Gene 3D
CDD
ProteinModelPortal
A0A2A4JDP2
A0A2H1X3Q8
A0A212FJL0
A0A194RPL9
A0A3S2P1T2
A0A0L7L8T4
+ More
A0A194PSD3 A0A1B6L590 A0A1B6JCU3 A0A1B6HVJ4 A0A0A9Z6H7 A0A0K8SA16 A0A146KTY3 A0A336LMI4 A0A1B6MGR6 E0VJ75 A0A1B6DB80 A0A182GFY5 A0A1W7R909 A0A023EJ25 A0A336MQQ6 A0A0N5DEJ7 B0WSF7 A0A1Y1N6P1 A0A2H8TK93 B4MBN6 F7EE13 A0A3B4BZI1 A0A3Q1GL66 A0A1B6M2D0 K7IVM0 J9FNI2 U3JZ48 A0A3P7EDL0 K7IVL8 A0A1B6E7K7 A0A1I8EAC3 A0A3B5B678 A0A336LIM0 A0A2S2P882 A0A2K8JLK0 A0A0R3RGP3 Q9VI09 Q1HQH6 A0A232F887 A0A1Q3FSX1 A0A1P6C0D3 A0A3Q3LS05 E3TGE4 R4WFB4 A0A2N9ISZ7 A0A0K8UQS4 A0A3Q0T5Y4 Q16QJ8 A0A0H5SCA5 C0SSN1 A0A218V388 A0A0N4SZJ6 A0A0H5SBL2 G7N5S0 M4CNJ5 A0A2P1BQH0 A0A1B1JEH8 A0A066WGK8 B5G191 B4PVQ0 B5G187 A0A3M0KFR0 J3MBF3 Q298A8 F5HPS3 A0A0S7MD46 A0A147B101 A0A3Q2QA16 A9LLZ5 A0A1A8ITN6 A0A1A8JIX3 A0A1A8NXD4 A0A1A8E8E3 A0A1A7Z8M9 Q8LSG9 A0A384L909 Q547P7 P93003 Q8LSH0 A0A182RZH2 R0HBP3 A0A1B1JEG7 A0A182ILC1 B0LQE2 B4NHB5 F4WAF3 A0A0Q9X5A3 B0LQE7 B4XEW2 B4G3B9 B4XEW0 A0A2P5X051
A0A194PSD3 A0A1B6L590 A0A1B6JCU3 A0A1B6HVJ4 A0A0A9Z6H7 A0A0K8SA16 A0A146KTY3 A0A336LMI4 A0A1B6MGR6 E0VJ75 A0A1B6DB80 A0A182GFY5 A0A1W7R909 A0A023EJ25 A0A336MQQ6 A0A0N5DEJ7 B0WSF7 A0A1Y1N6P1 A0A2H8TK93 B4MBN6 F7EE13 A0A3B4BZI1 A0A3Q1GL66 A0A1B6M2D0 K7IVM0 J9FNI2 U3JZ48 A0A3P7EDL0 K7IVL8 A0A1B6E7K7 A0A1I8EAC3 A0A3B5B678 A0A336LIM0 A0A2S2P882 A0A2K8JLK0 A0A0R3RGP3 Q9VI09 Q1HQH6 A0A232F887 A0A1Q3FSX1 A0A1P6C0D3 A0A3Q3LS05 E3TGE4 R4WFB4 A0A2N9ISZ7 A0A0K8UQS4 A0A3Q0T5Y4 Q16QJ8 A0A0H5SCA5 C0SSN1 A0A218V388 A0A0N4SZJ6 A0A0H5SBL2 G7N5S0 M4CNJ5 A0A2P1BQH0 A0A1B1JEH8 A0A066WGK8 B5G191 B4PVQ0 B5G187 A0A3M0KFR0 J3MBF3 Q298A8 F5HPS3 A0A0S7MD46 A0A147B101 A0A3Q2QA16 A9LLZ5 A0A1A8ITN6 A0A1A8JIX3 A0A1A8NXD4 A0A1A8E8E3 A0A1A7Z8M9 Q8LSG9 A0A384L909 Q547P7 P93003 Q8LSH0 A0A182RZH2 R0HBP3 A0A1B1JEG7 A0A182ILC1 B0LQE2 B4NHB5 F4WAF3 A0A0Q9X5A3 B0LQE7 B4XEW2 B4G3B9 B4XEW0 A0A2P5X051
PDB
5TVD
E-value=4.14719e-12,
Score=167
Ontologies
GO
PANTHER
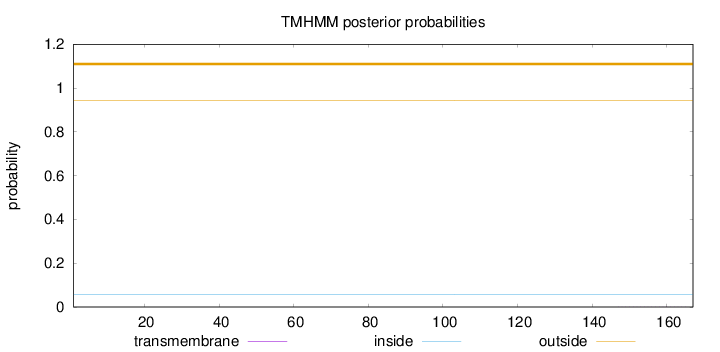
Topology
Subcellular location
Cytoplasm
Length:
167
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00651
Exp number, first 60 AAs:
0.00205
Total prob of N-in:
0.05584
outside
1 - 167
Population Genetic Test Statistics
Pi
207.506566
Theta
144.088144
Tajima's D
1.476298
CLR
0.133648
CSRT
0.779711014449277
Interpretation
Uncertain
