Gene
KWMTBOMO13729 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011500
Annotation
translation_initiation_factor_2_gamma_subunit_[Bombyx_mori]
Full name
Eukaryotic translation initiation factor 2 subunit 3
Alternative Name
Eukaryotic translation initiation factor 2 subunit gamma
Location in the cell
Mitochondrial Reliability : 2.25
Sequence
CDS
ATGGCTTCGAACGAAGGGCGAACTACTCAATCAAACTTGCATCAGCAAGACTTATCTAAATTGGATGTCACAAAATTATCTGCTCTCTCTCCCGAAGTCATATCAAGGCAGGCGACGATTAACATCGGTACAATTGGACATGTAGCTCATGGAAAATCTACTGTTGTGAAGGCTATTTCAGGAGTACAAACTGTAAGGTTTAAGAATGAATTGGAAAGAAATATTACGATCAAATTAGGTTATGCAAATGCAAAAATCTACCAATGCGACAATCCTAAATGTCCGCGGCCAACCAGCTTTATATCCGGCGGCTCATCTAAGGATGACAGCTTTCCGTGTCTTCGGCCTGCGTGTACGGGTCGCTTCCAACTAGTTCGCCACGTGAGCTTTGTAGACTGCCCTGGTCACGACATCCTTATGGCAACTATGCTTAACGGTGCTGCCGTGATGGACGCAGCATTACTACTCATTGCGGGCAACGAATCTTGCCCACAGCCCCAAACTAGTGAACACTTAGCAGCCATTGAGATAATGAAACTGAAGCATATCTTGATATTACAAAACAAGATCGATTTAGTTAAAGAGGGACAAGCTAAAGAACAGCATGAGCAAATTGTAAAATTCGTTCAAGGGACTGTGGCTGAAGGTGCACCTATTATACCCATATCGGCTCAACTCAAATATAATATTGAGGTCCTCTGCGAGTATATAACAAAAAAGATTCCCGTACCCCTAAGGGATTTCACATCGCCTCCTCGTATGATTGTTATTCGTTCATTCGATGTAAATAAGCCTGGTTGCGAGGTTGACGATCTACGGGGTGGCGTGGCAGGAGGATCAATTCTTCAAGGAGTTCTAACTGTCGGTATGGAAATCGAAGTGCGGCCTGGTTTAGTCAGTAAGGATGCAGATGGTAAATTAACTTGCCGACCCATATTTTCACGCATCGTTTCGCTGTTTGCCGAGCAGAATGAGCTACAGTATGCCGTCCCTGGTGGTCTTATTGGCGTAGGAACGAAGATCGAACCAACGCTGTGCCGTGCTGATAGACTTGTGGGACAAGTATTAGGAGCAGTTGGATGTTTACCGGGAATTTTTGTCAAACTCGAAGTATCTTATTATCTTTTGAAACGTTTGCTTGGCGTGCGCACTGAAGGAGATAAAAAGGCTGCCAAAGTCCAAAAATTAGTCAAAAATGAGGTCCTCCTCGTAAATATTGGATCGTTAAGTACTGGCGGCCGAGTAATCGCCACAAAAGTTGATTTGGCTAAAATCGCCCTTACGAACCCTGTTTGCACGGAAATTGGAGAAAAAGTAGCATTGAGTAGAAGAGTAGAAAATCACTGGCGGTTAATTGGTTGGGGACAAATTCAAGGAGGTACAACTATAGAACCAGCCAACAACTAA
Protein
MASNEGRTTQSNLHQQDLSKLDVTKLSALSPEVISRQATINIGTIGHVAHGKSTVVKAISGVQTVRFKNELERNITIKLGYANAKIYQCDNPKCPRPTSFISGGSSKDDSFPCLRPACTGRFQLVRHVSFVDCPGHDILMATMLNGAAVMDAALLLIAGNESCPQPQTSEHLAAIEIMKLKHILILQNKIDLVKEGQAKEQHEQIVKFVQGTVAEGAPIIPISAQLKYNIEVLCEYITKKIPVPLRDFTSPPRMIVIRSFDVNKPGCEVDDLRGGVAGGSILQGVLTVGMEIEVRPGLVSKDADGKLTCRPIFSRIVSLFAEQNELQYAVPGGLIGVGTKIEPTLCRADRLVGQVLGAVGCLPGIFVKLEVSYYLLKRLLGVRTEGDKKAAKVQKLVKNEVLLVNIGSLSTGGRVIATKVDLAKIALTNPVCTEIGEKVALSRRVENHWRLIGWGQIQGGTTIEPANN
Summary
Description
eIF-2 functions in the early steps of protein synthesis by forming a ternary complex with GTP and initiator tRNA. This complex binds to a 40S ribosomal subunit, followed by mRNA binding to form a 43S pre-initiation complex. Junction of the 60S ribosomal subunit to form the 80S initiation complex is preceded by hydrolysis of the GTP bound to eIF-2 and release of an eIF-2-GDP binary complex. In order for eIF-2 to recycle and catalyze another round of initiation, the GDP bound to eIF-2 must exchange with GTP by way of a reaction catalyzed by eIF-2B (By similarity).
Subunit
Heterotrimer composed of an alpha, a beta and a gamma chain.
Similarity
Belongs to the TRAFAC class translation factor GTPase superfamily. Classic translation factor GTPase family. EIF2G subfamily.
Keywords
Alternative splicing
Complete proteome
GTP-binding
Initiation factor
Nucleotide-binding
Protein biosynthesis
Reference proteome
Feature
chain Eukaryotic translation initiation factor 2 subunit 3
splice variant In isoform C.
splice variant In isoform C.
Uniprot
Q684K3
Q684K4
I4DJL2
S4P9E5
A0A0T6B140
Q9NAR5
+ More
A0A1W4X5V2 J3JYY1 A0A1B6H8G5 A0A1L8DRN2 Q9N9W0 A0A1Y1KJQ5 A0A232FFP6 K7IMQ9 Q684L1 Q684K5 T1DE69 A0A2A3EA31 A0A0C9RQZ7 A0A2M4A1J1 E0VWS1 A0A2M4CT05 A0A2M3YYY2 A0A2M4BM74 A0A158NT34 A0A182J245 F5HLB5 A0A026WVB1 A0A0K8TRR5 Q684K9 U5EUE9 A0A1B6C3T9 A0A1Q3FWU3 A0A023EV32 A0A0A9VXF7 Q684K7 A0A226EPT5 Q684L0 A0A0P4W0Q5 R4G8B0 A0A224XPH1 A0A0V0G300 A0A069DZ29 T1DE72 A0A0Q9X5F7 W8BYN8 A0A2S2QSH1 A0A1D2N4N4 A0A0Q9WG48 A0A0Q9X877 A0A2H8TQ64 A0A2S2PQ95 J9JTD7 A0A034VGU8 A0A0K8TYN3 A0A1L8EGX7 Q684L2 A0A3B0JNX4 I5APU1 A0A0R1E4U4 A0A1W4VRA7 A0A0B4KGP8 A0A0P8Y8M1 Q24208 B3P0T0 T1PDI5 A0A1I8PDI8 Q684K6 W4YX51 C3ZNT9 A0A1A9V2J9 E9GN88 A0A3Q3JXG2 A7SBR9 A0A0P5PBY7 A0A0P6BCZ9 Q24208-2 Q53XD3 A0A0N8E855 A0A3Q3B1G8 A0A0N8BSE6 A0A3Q3LVP7 A0A0N8A519 A0A3Q2FYI5 A0A0P4ZRB4 A0A0P5D865 Q4T3B4 A0A0P5SG74 A0A0P5L8B1 I3JNC5 H2S2P6 G1KGH9 D3TSF5 T1JPG6 A0A0P5I5P7 A0A3B3DVY7 A0A3S2NXS7 A0A0P5CYA9 A0A0P5FWZ2 A0A0P5MNQ3
A0A1W4X5V2 J3JYY1 A0A1B6H8G5 A0A1L8DRN2 Q9N9W0 A0A1Y1KJQ5 A0A232FFP6 K7IMQ9 Q684L1 Q684K5 T1DE69 A0A2A3EA31 A0A0C9RQZ7 A0A2M4A1J1 E0VWS1 A0A2M4CT05 A0A2M3YYY2 A0A2M4BM74 A0A158NT34 A0A182J245 F5HLB5 A0A026WVB1 A0A0K8TRR5 Q684K9 U5EUE9 A0A1B6C3T9 A0A1Q3FWU3 A0A023EV32 A0A0A9VXF7 Q684K7 A0A226EPT5 Q684L0 A0A0P4W0Q5 R4G8B0 A0A224XPH1 A0A0V0G300 A0A069DZ29 T1DE72 A0A0Q9X5F7 W8BYN8 A0A2S2QSH1 A0A1D2N4N4 A0A0Q9WG48 A0A0Q9X877 A0A2H8TQ64 A0A2S2PQ95 J9JTD7 A0A034VGU8 A0A0K8TYN3 A0A1L8EGX7 Q684L2 A0A3B0JNX4 I5APU1 A0A0R1E4U4 A0A1W4VRA7 A0A0B4KGP8 A0A0P8Y8M1 Q24208 B3P0T0 T1PDI5 A0A1I8PDI8 Q684K6 W4YX51 C3ZNT9 A0A1A9V2J9 E9GN88 A0A3Q3JXG2 A7SBR9 A0A0P5PBY7 A0A0P6BCZ9 Q24208-2 Q53XD3 A0A0N8E855 A0A3Q3B1G8 A0A0N8BSE6 A0A3Q3LVP7 A0A0N8A519 A0A3Q2FYI5 A0A0P4ZRB4 A0A0P5D865 Q4T3B4 A0A0P5SG74 A0A0P5L8B1 I3JNC5 H2S2P6 G1KGH9 D3TSF5 T1JPG6 A0A0P5I5P7 A0A3B3DVY7 A0A3S2NXS7 A0A0P5CYA9 A0A0P5FWZ2 A0A0P5MNQ3
Pubmed
15356279
22651552
23622113
11063691
22516182
28004739
+ More
28648823 20075255 24330624 20566863 21347285 12364791 14747013 17210077 24508170 26369729 24945155 25401762 26823975 27129103 26334808 17994087 24495485 27289101 25348373 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 7915232 12537569 25315136 18563158 21292972 17615350 15496914 25186727 21551351 20353571 29451363
28648823 20075255 24330624 20566863 21347285 12364791 14747013 17210077 24508170 26369729 24945155 25401762 26823975 27129103 26334808 17994087 24495485 27289101 25348373 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 7915232 12537569 25315136 18563158 21292972 17615350 15496914 25186727 21551351 20353571 29451363
EMBL
AJ715870
CAG29676.1
AJ715869
CAG29675.1
AK401480
BAM18102.1
+ More
GAIX01005526 JAA87034.1 LJIG01016274 KRT81094.1 AJ290961 AJ290962 AJ290963 CAB98197.1 BT128462 AEE63419.1 GECU01036718 JAS70988.1 GFDF01004985 JAV09099.1 AJ290964 CAB94834.1 GEZM01081401 JAV61649.1 NNAY01000314 OXU29278.1 AJ715862 CAG29668.1 AJ715868 CAG29674.1 GALA01001162 JAA93690.1 KZ288325 PBC28036.1 GBYB01009701 JAG79468.1 GGFK01001298 MBW34619.1 DS235824 EEB17827.1 GGFL01004272 MBW68450.1 GGFM01000715 MBW21466.1 GGFJ01004953 MBW54094.1 ADTU01025481 AAAB01008888 EGK97076.1 KK107109 EZA59044.1 GDAI01000544 JAI17059.1 AJ715864 CAG29670.1 GANO01002355 JAB57516.1 GEDC01029398 JAS07900.1 GFDL01003152 JAV31893.1 GAPW01001259 JAC12339.1 GBHO01042717 GBRD01008334 GDHC01018866 JAG00887.1 JAG57487.1 JAP99762.1 AJ715866 CAG29672.1 LNIX01000003 OXA58596.1 AJ715863 CAG29669.1 GDKW01001313 JAI55282.1 ACPB03002356 GAHY01001287 JAA76223.1 GFTR01006505 JAW09921.1 GECL01003625 JAP02499.1 GBGD01001360 JAC87529.1 GALA01001157 JAA93695.1 CH964232 KRF99458.1 GAMC01004787 JAC01769.1 GGMS01011257 MBY80460.1 LJIJ01000221 ODN00237.1 CH940650 KRF83186.1 CH933806 KRG01428.1 GFXV01004532 MBW16337.1 GGMR01018397 MBY31016.1 ABLF02034802 ABLF02034812 GAKP01018189 JAC40763.1 GDHF01032941 JAI19373.1 GFDG01000832 JAV17967.1 AJ715861 CAG29667.1 OUUW01000008 SPP83934.1 CM000070 EIM52976.1 CM000160 KRK03774.1 AE014297 AGB95968.1 CH902629 KPU75544.1 X80069 AJ290956 AY061102 CH954181 EDV48978.1 KA646195 AFP60824.1 AJ715867 CAG29673.1 AAGJ04152272 GG666653 EEN45821.1 GL732554 EFX79075.1 DS469618 EDO38840.1 GDIQ01140287 JAL11439.1 GDIP01023404 LRGB01003257 JAM80311.1 KZS03523.1 BT012446 AAS93717.1 GDIQ01052428 JAN42309.1 GDIQ01149491 JAL02235.1 GDIP01181785 JAJ41617.1 GDIP01209775 JAJ13627.1 GDIP01161533 JAJ61869.1 CAAE01010102 CAF92618.1 GDIP01140454 JAL63260.1 GDIQ01178712 JAK73013.1 AERX01040597 AERX01040598 EZ424357 ADD20633.1 JH431826 GDIQ01219055 GDIQ01219054 JAK32670.1 CM012456 RVE59153.1 GDIP01179982 JAJ43420.1 GDIQ01250504 JAK01221.1 GDIQ01176556 JAK75169.1
GAIX01005526 JAA87034.1 LJIG01016274 KRT81094.1 AJ290961 AJ290962 AJ290963 CAB98197.1 BT128462 AEE63419.1 GECU01036718 JAS70988.1 GFDF01004985 JAV09099.1 AJ290964 CAB94834.1 GEZM01081401 JAV61649.1 NNAY01000314 OXU29278.1 AJ715862 CAG29668.1 AJ715868 CAG29674.1 GALA01001162 JAA93690.1 KZ288325 PBC28036.1 GBYB01009701 JAG79468.1 GGFK01001298 MBW34619.1 DS235824 EEB17827.1 GGFL01004272 MBW68450.1 GGFM01000715 MBW21466.1 GGFJ01004953 MBW54094.1 ADTU01025481 AAAB01008888 EGK97076.1 KK107109 EZA59044.1 GDAI01000544 JAI17059.1 AJ715864 CAG29670.1 GANO01002355 JAB57516.1 GEDC01029398 JAS07900.1 GFDL01003152 JAV31893.1 GAPW01001259 JAC12339.1 GBHO01042717 GBRD01008334 GDHC01018866 JAG00887.1 JAG57487.1 JAP99762.1 AJ715866 CAG29672.1 LNIX01000003 OXA58596.1 AJ715863 CAG29669.1 GDKW01001313 JAI55282.1 ACPB03002356 GAHY01001287 JAA76223.1 GFTR01006505 JAW09921.1 GECL01003625 JAP02499.1 GBGD01001360 JAC87529.1 GALA01001157 JAA93695.1 CH964232 KRF99458.1 GAMC01004787 JAC01769.1 GGMS01011257 MBY80460.1 LJIJ01000221 ODN00237.1 CH940650 KRF83186.1 CH933806 KRG01428.1 GFXV01004532 MBW16337.1 GGMR01018397 MBY31016.1 ABLF02034802 ABLF02034812 GAKP01018189 JAC40763.1 GDHF01032941 JAI19373.1 GFDG01000832 JAV17967.1 AJ715861 CAG29667.1 OUUW01000008 SPP83934.1 CM000070 EIM52976.1 CM000160 KRK03774.1 AE014297 AGB95968.1 CH902629 KPU75544.1 X80069 AJ290956 AY061102 CH954181 EDV48978.1 KA646195 AFP60824.1 AJ715867 CAG29673.1 AAGJ04152272 GG666653 EEN45821.1 GL732554 EFX79075.1 DS469618 EDO38840.1 GDIQ01140287 JAL11439.1 GDIP01023404 LRGB01003257 JAM80311.1 KZS03523.1 BT012446 AAS93717.1 GDIQ01052428 JAN42309.1 GDIQ01149491 JAL02235.1 GDIP01181785 JAJ41617.1 GDIP01209775 JAJ13627.1 GDIP01161533 JAJ61869.1 CAAE01010102 CAF92618.1 GDIP01140454 JAL63260.1 GDIQ01178712 JAK73013.1 AERX01040597 AERX01040598 EZ424357 ADD20633.1 JH431826 GDIQ01219055 GDIQ01219054 JAK32670.1 CM012456 RVE59153.1 GDIP01179982 JAJ43420.1 GDIQ01250504 JAK01221.1 GDIQ01176556 JAK75169.1
Proteomes
UP000192223
UP000215335
UP000002358
UP000242457
UP000009046
UP000005205
+ More
UP000075880 UP000007062 UP000053097 UP000198287 UP000015103 UP000007798 UP000094527 UP000008792 UP000009192 UP000007819 UP000268350 UP000001819 UP000002282 UP000192221 UP000000803 UP000007801 UP000008711 UP000095301 UP000095300 UP000007110 UP000001554 UP000078200 UP000000305 UP000261600 UP000001593 UP000076858 UP000264800 UP000261640 UP000265020 UP000007303 UP000005207 UP000005226 UP000001646 UP000261560
UP000075880 UP000007062 UP000053097 UP000198287 UP000015103 UP000007798 UP000094527 UP000008792 UP000009192 UP000007819 UP000268350 UP000001819 UP000002282 UP000192221 UP000000803 UP000007801 UP000008711 UP000095301 UP000095300 UP000007110 UP000001554 UP000078200 UP000000305 UP000261600 UP000001593 UP000076858 UP000264800 UP000261640 UP000265020 UP000007303 UP000005207 UP000005226 UP000001646 UP000261560
Interpro
Gene 3D
ProteinModelPortal
Q684K3
Q684K4
I4DJL2
S4P9E5
A0A0T6B140
Q9NAR5
+ More
A0A1W4X5V2 J3JYY1 A0A1B6H8G5 A0A1L8DRN2 Q9N9W0 A0A1Y1KJQ5 A0A232FFP6 K7IMQ9 Q684L1 Q684K5 T1DE69 A0A2A3EA31 A0A0C9RQZ7 A0A2M4A1J1 E0VWS1 A0A2M4CT05 A0A2M3YYY2 A0A2M4BM74 A0A158NT34 A0A182J245 F5HLB5 A0A026WVB1 A0A0K8TRR5 Q684K9 U5EUE9 A0A1B6C3T9 A0A1Q3FWU3 A0A023EV32 A0A0A9VXF7 Q684K7 A0A226EPT5 Q684L0 A0A0P4W0Q5 R4G8B0 A0A224XPH1 A0A0V0G300 A0A069DZ29 T1DE72 A0A0Q9X5F7 W8BYN8 A0A2S2QSH1 A0A1D2N4N4 A0A0Q9WG48 A0A0Q9X877 A0A2H8TQ64 A0A2S2PQ95 J9JTD7 A0A034VGU8 A0A0K8TYN3 A0A1L8EGX7 Q684L2 A0A3B0JNX4 I5APU1 A0A0R1E4U4 A0A1W4VRA7 A0A0B4KGP8 A0A0P8Y8M1 Q24208 B3P0T0 T1PDI5 A0A1I8PDI8 Q684K6 W4YX51 C3ZNT9 A0A1A9V2J9 E9GN88 A0A3Q3JXG2 A7SBR9 A0A0P5PBY7 A0A0P6BCZ9 Q24208-2 Q53XD3 A0A0N8E855 A0A3Q3B1G8 A0A0N8BSE6 A0A3Q3LVP7 A0A0N8A519 A0A3Q2FYI5 A0A0P4ZRB4 A0A0P5D865 Q4T3B4 A0A0P5SG74 A0A0P5L8B1 I3JNC5 H2S2P6 G1KGH9 D3TSF5 T1JPG6 A0A0P5I5P7 A0A3B3DVY7 A0A3S2NXS7 A0A0P5CYA9 A0A0P5FWZ2 A0A0P5MNQ3
A0A1W4X5V2 J3JYY1 A0A1B6H8G5 A0A1L8DRN2 Q9N9W0 A0A1Y1KJQ5 A0A232FFP6 K7IMQ9 Q684L1 Q684K5 T1DE69 A0A2A3EA31 A0A0C9RQZ7 A0A2M4A1J1 E0VWS1 A0A2M4CT05 A0A2M3YYY2 A0A2M4BM74 A0A158NT34 A0A182J245 F5HLB5 A0A026WVB1 A0A0K8TRR5 Q684K9 U5EUE9 A0A1B6C3T9 A0A1Q3FWU3 A0A023EV32 A0A0A9VXF7 Q684K7 A0A226EPT5 Q684L0 A0A0P4W0Q5 R4G8B0 A0A224XPH1 A0A0V0G300 A0A069DZ29 T1DE72 A0A0Q9X5F7 W8BYN8 A0A2S2QSH1 A0A1D2N4N4 A0A0Q9WG48 A0A0Q9X877 A0A2H8TQ64 A0A2S2PQ95 J9JTD7 A0A034VGU8 A0A0K8TYN3 A0A1L8EGX7 Q684L2 A0A3B0JNX4 I5APU1 A0A0R1E4U4 A0A1W4VRA7 A0A0B4KGP8 A0A0P8Y8M1 Q24208 B3P0T0 T1PDI5 A0A1I8PDI8 Q684K6 W4YX51 C3ZNT9 A0A1A9V2J9 E9GN88 A0A3Q3JXG2 A7SBR9 A0A0P5PBY7 A0A0P6BCZ9 Q24208-2 Q53XD3 A0A0N8E855 A0A3Q3B1G8 A0A0N8BSE6 A0A3Q3LVP7 A0A0N8A519 A0A3Q2FYI5 A0A0P4ZRB4 A0A0P5D865 Q4T3B4 A0A0P5SG74 A0A0P5L8B1 I3JNC5 H2S2P6 G1KGH9 D3TSF5 T1JPG6 A0A0P5I5P7 A0A3B3DVY7 A0A3S2NXS7 A0A0P5CYA9 A0A0P5FWZ2 A0A0P5MNQ3
PDB
6O85
E-value=0,
Score=1918
Ontologies
GO
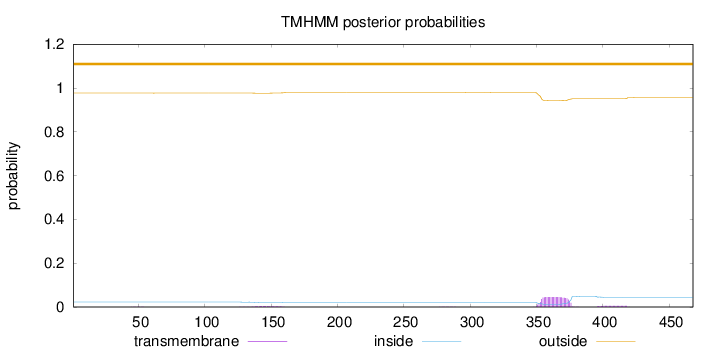
Topology
Length:
468
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.21156
Exp number, first 60 AAs:
0.00279
Total prob of N-in:
0.02302
outside
1 - 468
Population Genetic Test Statistics
Pi
2.852734
Theta
183.169328
Tajima's D
-2.447287
CLR
207.404934
CSRT
0.000899955002249887
Interpretation
Uncertain
