Gene
KWMTBOMO13725
Pre Gene Modal
BGIBMGA011503
Annotation
PREDICTED:_peroxisomal_N(1)-acetyl-spermine/spermidine_oxidase_isoform_X1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.932 Nuclear Reliability : 1.947
Sequence
CDS
ATGGGTGACGAAAATTTAGGTTCCAGTGAAAAATGTAATAGTTCCAAAAAGTTTAAAGTCATTATCATTGGCTGTGGAATAGCTGGCTTATCCTCAGCCAACTATTTGCTTCAAAATGGCATGGAAGATTTCTTAATATTAGAAGCAAGAAAACGTATCGGTGGACGCATAATATCTATACCAATGAAGAGTCATAATGTTGAGCTCGGTGCTAATTGGATTCATGGTGTTTTAGGAAATCCTATCTTTGAATTAGCAATGTCCCACAATCTAGTAAATATAATAAATATACCGAAGCCCCATAAAGTTATAGCAGCTACAGAAAATGGCAAGCAAGTGCCGTTTGGTATTCTGCATGAAATACATGAGGCCTACGTATGTTTTCTTAGGCGATGCGAAGAGTATTTCTTGTGTCAATATTTACCTCCCCCAGATATACACAGCGTTGGAGAGCATATAAATTTAGAAGCAACTTTATATTTGGAGCGCTTGCCATCTTCTGAGGAAAAGAAGCTAAGAAGACTAATATTTGACTGTTTGTTAAAACGTGAAACCTGCATATCAGGTTGCCACAGTATGGATGAAGTAGATCTTCTTGAATTAGGTAGTTATACTGAACTTCAAGGAGGTAATATTATGATACCTTCAGGTTTTAGTTCTATTTTAGACCCATTAAAGAAAAACATAAATGATAAAAAAATATTGACAAACCATGCGGTGAGGAAAATTATATGGGATTCTGATGTAAACCAATCACTGAAATCACTTGATGATACAGGTGAGGAATCTGAAGATTCTGATCAAACTGTGATAGAAGACAACTCTAAAATTTCACCTGACAAATCATTAAAACCTAATATAGAAAACTCTTTACAAGATATGACTCAACAACCAAAGAAAAAGTACAATAATTATGTTGAGATTGTGTGCGAAAATGGACAATCATTTCATGCAAATCATGTAATTTGTACTATTCCACTAGGTGTTCTCAAAGAGAAAGCATCAAGTTTATTTGAACCAAAGCTGCCATCATACAAAATAGAATCAATTGAAAAATTATTATTTGGTGTTGTAGATAAAATATACTTAGAGTATGAGAGACCCTTTTTAAACCCAGATATAACGGAAATAATGCTTTTGTGGGAAACTCCTACTACACCTGAAGATATGTCTCAGTCTTGGTACAAAAAAATTTACTCATTTAGTAAAGTATCAGAAACTCTCTTATTAGGATGGGTTTCTGGTAAAGAAGCAGAGTATATGGAAACATTGTCTATGGAAGAGGTGGCAGAAACTTGTACAAAAATATTACGAAAATTTCTGAATGACCCTTTTGTACCGGAACCTCAAAAATGTGTTTGTACCAGTTGGAGAGAACAGCCCTATACGAGAGGATCATATACTGCAATTGCTGTTGGAGCCAGTCAAAGTGATGTAGAGAGTCTTGCTCAACCAATCTTTAGAAATGTACACGATAAGAAGCCTGTTCTACTCTTTGCCGGAGAGCATACACACAGCAGCTATTATTCTACGGCACATGGTGCTTATTTATCAGGTCAAATAGCAGCTCGAAGACTTCTAGCACCTGATGAAGCGTCAGAAGGCACACTTGACTGTCCAGATTCAGCGGATCTCAATTCTTGGATACAAGGAATACAGCTGGAAGGGTGA
Protein
MGDENLGSSEKCNSSKKFKVIIIGCGIAGLSSANYLLQNGMEDFLILEARKRIGGRIISIPMKSHNVELGANWIHGVLGNPIFELAMSHNLVNIINIPKPHKVIAATENGKQVPFGILHEIHEAYVCFLRRCEEYFLCQYLPPPDIHSVGEHINLEATLYLERLPSSEEKKLRRLIFDCLLKRETCISGCHSMDEVDLLELGSYTELQGGNIMIPSGFSSILDPLKKNINDKKILTNHAVRKIIWDSDVNQSLKSLDDTGEESEDSDQTVIEDNSKISPDKSLKPNIENSLQDMTQQPKKKYNNYVEIVCENGQSFHANHVICTIPLGVLKEKASSLFEPKLPSYKIESIEKLLFGVVDKIYLEYERPFLNPDITEIMLLWETPTTPEDMSQSWYKKIYSFSKVSETLLLGWVSGKEAEYMETLSMEEVAETCTKILRKFLNDPFVPEPQKCVCTSWREQPYTRGSYTAIAVGASQSDVESLAQPIFRNVHDKKPVLLFAGEHTHSSYYSTAHGAYLSGQIAARRLLAPDEASEGTLDCPDSADLNSWIQGIQLEG
Summary
Uniprot
H9JPP8
A0A2H1VHI5
A0A2A4JA60
A0A194PSD8
A0A194RPL4
A0A0L7L6U0
+ More
A0A212FJM2 A0A3S2TFU4 A0A084WTN2 A0A1B6MI67 A0A182PI53 A0A182LEI5 A0A182W0U2 A0A182Y6K5 A0A182RXR3 A0A182M237 A0A182NRF1 A0A182Q5F4 A0A182HSH9 Q7QCG5 A0A182TSW5 A0A182VJG9 A0A182IKS7 A0A1B6EDQ6 A0A182XAN9 A0A1W4X3D2 A0A182STX1 T1HND8 Q171T9 A0A1S4FGT1 A0A067RHU3 A0A023F333 A0A182G0L6 A0A2M4ATP0 A0A0L0BWQ9 W5JV05 A0A182FAZ2 A0A1W4W229 B4KCP3 Q297N9 B4PT83 B4M502 A0A2J7Q4K7 A0A3B0JRU4 B4G2T3 B4JSR9 Q9VHN8 B3M234 B4QY00 A0A1A9Z860 A0A1A9VDG6 B4HL37 A0A0A1X338 A0A1A9XM60 B3P4V1 A0A1Y1KUI6 T1PJA2 A0A1B0BL06 D6WAZ4 A0A0K8U049 A0A1I8Q714 B4NLA6 N6TR36 W8B5G7 A0A0M3QXB7 A0A034VXM1 U4U2C9 A0A182K4J9 A0A0A9WZT7 A0A336MCA5 A0A2P8ZMW8 B0XF78 E2A7L3 A0A154PTR4 A0A0L7QU48 A0A088A1V9 A0A2A3ELJ3 F4X262 A0A0M9A0W8 A0A1B0DA32 A0A195CK40 A0A195DQI8 E2BPK1 A0A151X5K3 A0A1W4WTU5 A0A026WZG3 A0A151I614 A0A195EWH2 A0A158NAJ3 A0A2M4AUB9 K7JA85 A0A069DTR7 A0A2R7WPL3 A0A1B0FDY0 A0A1A9WYS5 A0A1Y1KPM3 A0A182GCL4 E9IF49 A0A0R3NN60 A0A0N7ZCF4
A0A212FJM2 A0A3S2TFU4 A0A084WTN2 A0A1B6MI67 A0A182PI53 A0A182LEI5 A0A182W0U2 A0A182Y6K5 A0A182RXR3 A0A182M237 A0A182NRF1 A0A182Q5F4 A0A182HSH9 Q7QCG5 A0A182TSW5 A0A182VJG9 A0A182IKS7 A0A1B6EDQ6 A0A182XAN9 A0A1W4X3D2 A0A182STX1 T1HND8 Q171T9 A0A1S4FGT1 A0A067RHU3 A0A023F333 A0A182G0L6 A0A2M4ATP0 A0A0L0BWQ9 W5JV05 A0A182FAZ2 A0A1W4W229 B4KCP3 Q297N9 B4PT83 B4M502 A0A2J7Q4K7 A0A3B0JRU4 B4G2T3 B4JSR9 Q9VHN8 B3M234 B4QY00 A0A1A9Z860 A0A1A9VDG6 B4HL37 A0A0A1X338 A0A1A9XM60 B3P4V1 A0A1Y1KUI6 T1PJA2 A0A1B0BL06 D6WAZ4 A0A0K8U049 A0A1I8Q714 B4NLA6 N6TR36 W8B5G7 A0A0M3QXB7 A0A034VXM1 U4U2C9 A0A182K4J9 A0A0A9WZT7 A0A336MCA5 A0A2P8ZMW8 B0XF78 E2A7L3 A0A154PTR4 A0A0L7QU48 A0A088A1V9 A0A2A3ELJ3 F4X262 A0A0M9A0W8 A0A1B0DA32 A0A195CK40 A0A195DQI8 E2BPK1 A0A151X5K3 A0A1W4WTU5 A0A026WZG3 A0A151I614 A0A195EWH2 A0A158NAJ3 A0A2M4AUB9 K7JA85 A0A069DTR7 A0A2R7WPL3 A0A1B0FDY0 A0A1A9WYS5 A0A1Y1KPM3 A0A182GCL4 E9IF49 A0A0R3NN60 A0A0N7ZCF4
Pubmed
19121390
26354079
26227816
22118469
24438588
20966253
+ More
25244985 12364791 14747013 17210077 17510324 24845553 25474469 26483478 26108605 20920257 23761445 17994087 15632085 23185243 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 28004739 25315136 18362917 19820115 23537049 24495485 25348373 25401762 26823975 29403074 20798317 21719571 24508170 30249741 21347285 20075255 26334808 21282665
25244985 12364791 14747013 17210077 17510324 24845553 25474469 26483478 26108605 20920257 23761445 17994087 15632085 23185243 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 28004739 25315136 18362917 19820115 23537049 24495485 25348373 25401762 26823975 29403074 20798317 21719571 24508170 30249741 21347285 20075255 26334808 21282665
EMBL
BABH01015150
ODYU01002594
SOQ40303.1
NWSH01002449
PCG68283.1
KQ459601
+ More
KPI94040.1 KQ459833 KPJ19793.1 JTDY01002630 KOB71064.1 AGBW02008253 OWR53931.1 RSAL01000184 RVE44839.1 ATLV01026902 KE525420 KFB53576.1 GEBQ01004329 JAT35648.1 AXCM01002172 AXCN02001464 APCN01000279 AAAB01008859 EAA08089.4 GEDC01001219 JAS36079.1 ACPB03001835 CH477446 EAT40769.1 KK852672 KDR18787.1 GBBI01003181 JAC15531.1 JXUM01134760 KQ568171 KXJ69112.1 GGFK01010751 MBW44072.1 JRES01001227 KNC24463.1 ADMH02000285 ETN67118.1 CH933806 EDW15892.1 CM000070 EAL28166.1 KRT00516.1 CM000160 EDW96544.1 CH940652 EDW59713.2 NEVH01018378 PNF23513.1 OUUW01000007 SPP83112.1 CH479179 EDW24128.1 CH916373 EDV94809.1 AE014297 AY069611 AAF54264.1 AAL39756.1 CH902617 EDV43358.1 CM000364 EDX13737.1 CH480815 EDW43001.1 GBXI01009194 JAD05098.1 CH954181 EDV49895.1 GEZM01079044 GEZM01079041 JAV62527.1 KA648200 AFP62829.1 JXJN01016136 KQ971311 EEZ98988.1 GDHF01032416 JAI19898.1 CH964272 EDW84309.1 APGK01055062 KB741259 ENN71700.1 GAMC01018032 GAMC01018031 GAMC01018030 GAMC01018029 JAB88526.1 CP012526 ALC45643.1 GAKP01012684 JAC46268.1 KB631924 ERL87222.1 GBHO01031586 GBHO01031581 GBHO01001446 GDHC01020982 JAG12018.1 JAG12023.1 JAG42158.1 JAP97646.1 UFQS01000920 UFQT01000920 SSX07694.1 SSX28022.1 PYGN01000014 PSN57820.1 DS232906 EDS26544.1 GL437348 EFN70585.1 KQ435127 KZC14824.1 KQ414735 KOC62187.1 KZ288215 PBC32673.1 GL888565 EGI59478.1 KQ435776 KOX74897.1 AJVK01028749 KQ977642 KYN01100.1 KQ980612 KYN15128.1 GL449633 EFN82405.1 KQ982519 KYQ55548.1 KK107077 QOIP01000010 EZA60534.1 RLU17351.1 KQ976413 KYM90397.1 KQ981954 KYN32234.1 ADTU01010339 GGFK01011056 MBW44377.1 AAZX01006344 GBGD01001416 JAC87473.1 KK855026 PTY20375.1 CCAG010002891 GEZM01079043 JAV62528.1 JXUM01009655 KQ560288 KXJ83244.1 GL762758 EFZ20860.1 KRT00515.1 GDRN01067909 JAI64296.1
KPI94040.1 KQ459833 KPJ19793.1 JTDY01002630 KOB71064.1 AGBW02008253 OWR53931.1 RSAL01000184 RVE44839.1 ATLV01026902 KE525420 KFB53576.1 GEBQ01004329 JAT35648.1 AXCM01002172 AXCN02001464 APCN01000279 AAAB01008859 EAA08089.4 GEDC01001219 JAS36079.1 ACPB03001835 CH477446 EAT40769.1 KK852672 KDR18787.1 GBBI01003181 JAC15531.1 JXUM01134760 KQ568171 KXJ69112.1 GGFK01010751 MBW44072.1 JRES01001227 KNC24463.1 ADMH02000285 ETN67118.1 CH933806 EDW15892.1 CM000070 EAL28166.1 KRT00516.1 CM000160 EDW96544.1 CH940652 EDW59713.2 NEVH01018378 PNF23513.1 OUUW01000007 SPP83112.1 CH479179 EDW24128.1 CH916373 EDV94809.1 AE014297 AY069611 AAF54264.1 AAL39756.1 CH902617 EDV43358.1 CM000364 EDX13737.1 CH480815 EDW43001.1 GBXI01009194 JAD05098.1 CH954181 EDV49895.1 GEZM01079044 GEZM01079041 JAV62527.1 KA648200 AFP62829.1 JXJN01016136 KQ971311 EEZ98988.1 GDHF01032416 JAI19898.1 CH964272 EDW84309.1 APGK01055062 KB741259 ENN71700.1 GAMC01018032 GAMC01018031 GAMC01018030 GAMC01018029 JAB88526.1 CP012526 ALC45643.1 GAKP01012684 JAC46268.1 KB631924 ERL87222.1 GBHO01031586 GBHO01031581 GBHO01001446 GDHC01020982 JAG12018.1 JAG12023.1 JAG42158.1 JAP97646.1 UFQS01000920 UFQT01000920 SSX07694.1 SSX28022.1 PYGN01000014 PSN57820.1 DS232906 EDS26544.1 GL437348 EFN70585.1 KQ435127 KZC14824.1 KQ414735 KOC62187.1 KZ288215 PBC32673.1 GL888565 EGI59478.1 KQ435776 KOX74897.1 AJVK01028749 KQ977642 KYN01100.1 KQ980612 KYN15128.1 GL449633 EFN82405.1 KQ982519 KYQ55548.1 KK107077 QOIP01000010 EZA60534.1 RLU17351.1 KQ976413 KYM90397.1 KQ981954 KYN32234.1 ADTU01010339 GGFK01011056 MBW44377.1 AAZX01006344 GBGD01001416 JAC87473.1 KK855026 PTY20375.1 CCAG010002891 GEZM01079043 JAV62528.1 JXUM01009655 KQ560288 KXJ83244.1 GL762758 EFZ20860.1 KRT00515.1 GDRN01067909 JAI64296.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000037510
UP000007151
+ More
UP000283053 UP000030765 UP000075885 UP000075882 UP000075920 UP000076408 UP000075900 UP000075883 UP000075884 UP000075886 UP000075840 UP000007062 UP000075902 UP000075903 UP000075880 UP000076407 UP000192223 UP000075901 UP000015103 UP000008820 UP000027135 UP000069940 UP000249989 UP000037069 UP000000673 UP000069272 UP000192221 UP000009192 UP000001819 UP000002282 UP000008792 UP000235965 UP000268350 UP000008744 UP000001070 UP000000803 UP000007801 UP000000304 UP000092445 UP000078200 UP000001292 UP000092443 UP000008711 UP000095301 UP000092460 UP000007266 UP000095300 UP000007798 UP000019118 UP000092553 UP000030742 UP000075881 UP000245037 UP000002320 UP000000311 UP000076502 UP000053825 UP000005203 UP000242457 UP000007755 UP000053105 UP000092462 UP000078542 UP000078492 UP000008237 UP000075809 UP000053097 UP000279307 UP000078540 UP000078541 UP000005205 UP000002358 UP000092444 UP000091820
UP000283053 UP000030765 UP000075885 UP000075882 UP000075920 UP000076408 UP000075900 UP000075883 UP000075884 UP000075886 UP000075840 UP000007062 UP000075902 UP000075903 UP000075880 UP000076407 UP000192223 UP000075901 UP000015103 UP000008820 UP000027135 UP000069940 UP000249989 UP000037069 UP000000673 UP000069272 UP000192221 UP000009192 UP000001819 UP000002282 UP000008792 UP000235965 UP000268350 UP000008744 UP000001070 UP000000803 UP000007801 UP000000304 UP000092445 UP000078200 UP000001292 UP000092443 UP000008711 UP000095301 UP000092460 UP000007266 UP000095300 UP000007798 UP000019118 UP000092553 UP000030742 UP000075881 UP000245037 UP000002320 UP000000311 UP000076502 UP000053825 UP000005203 UP000242457 UP000007755 UP000053105 UP000092462 UP000078542 UP000078492 UP000008237 UP000075809 UP000053097 UP000279307 UP000078540 UP000078541 UP000005205 UP000002358 UP000092444 UP000091820
Pfam
PF01593 Amino_oxidase
SUPFAM
SSF51905
SSF51905
Gene 3D
ProteinModelPortal
H9JPP8
A0A2H1VHI5
A0A2A4JA60
A0A194PSD8
A0A194RPL4
A0A0L7L6U0
+ More
A0A212FJM2 A0A3S2TFU4 A0A084WTN2 A0A1B6MI67 A0A182PI53 A0A182LEI5 A0A182W0U2 A0A182Y6K5 A0A182RXR3 A0A182M237 A0A182NRF1 A0A182Q5F4 A0A182HSH9 Q7QCG5 A0A182TSW5 A0A182VJG9 A0A182IKS7 A0A1B6EDQ6 A0A182XAN9 A0A1W4X3D2 A0A182STX1 T1HND8 Q171T9 A0A1S4FGT1 A0A067RHU3 A0A023F333 A0A182G0L6 A0A2M4ATP0 A0A0L0BWQ9 W5JV05 A0A182FAZ2 A0A1W4W229 B4KCP3 Q297N9 B4PT83 B4M502 A0A2J7Q4K7 A0A3B0JRU4 B4G2T3 B4JSR9 Q9VHN8 B3M234 B4QY00 A0A1A9Z860 A0A1A9VDG6 B4HL37 A0A0A1X338 A0A1A9XM60 B3P4V1 A0A1Y1KUI6 T1PJA2 A0A1B0BL06 D6WAZ4 A0A0K8U049 A0A1I8Q714 B4NLA6 N6TR36 W8B5G7 A0A0M3QXB7 A0A034VXM1 U4U2C9 A0A182K4J9 A0A0A9WZT7 A0A336MCA5 A0A2P8ZMW8 B0XF78 E2A7L3 A0A154PTR4 A0A0L7QU48 A0A088A1V9 A0A2A3ELJ3 F4X262 A0A0M9A0W8 A0A1B0DA32 A0A195CK40 A0A195DQI8 E2BPK1 A0A151X5K3 A0A1W4WTU5 A0A026WZG3 A0A151I614 A0A195EWH2 A0A158NAJ3 A0A2M4AUB9 K7JA85 A0A069DTR7 A0A2R7WPL3 A0A1B0FDY0 A0A1A9WYS5 A0A1Y1KPM3 A0A182GCL4 E9IF49 A0A0R3NN60 A0A0N7ZCF4
A0A212FJM2 A0A3S2TFU4 A0A084WTN2 A0A1B6MI67 A0A182PI53 A0A182LEI5 A0A182W0U2 A0A182Y6K5 A0A182RXR3 A0A182M237 A0A182NRF1 A0A182Q5F4 A0A182HSH9 Q7QCG5 A0A182TSW5 A0A182VJG9 A0A182IKS7 A0A1B6EDQ6 A0A182XAN9 A0A1W4X3D2 A0A182STX1 T1HND8 Q171T9 A0A1S4FGT1 A0A067RHU3 A0A023F333 A0A182G0L6 A0A2M4ATP0 A0A0L0BWQ9 W5JV05 A0A182FAZ2 A0A1W4W229 B4KCP3 Q297N9 B4PT83 B4M502 A0A2J7Q4K7 A0A3B0JRU4 B4G2T3 B4JSR9 Q9VHN8 B3M234 B4QY00 A0A1A9Z860 A0A1A9VDG6 B4HL37 A0A0A1X338 A0A1A9XM60 B3P4V1 A0A1Y1KUI6 T1PJA2 A0A1B0BL06 D6WAZ4 A0A0K8U049 A0A1I8Q714 B4NLA6 N6TR36 W8B5G7 A0A0M3QXB7 A0A034VXM1 U4U2C9 A0A182K4J9 A0A0A9WZT7 A0A336MCA5 A0A2P8ZMW8 B0XF78 E2A7L3 A0A154PTR4 A0A0L7QU48 A0A088A1V9 A0A2A3ELJ3 F4X262 A0A0M9A0W8 A0A1B0DA32 A0A195CK40 A0A195DQI8 E2BPK1 A0A151X5K3 A0A1W4WTU5 A0A026WZG3 A0A151I614 A0A195EWH2 A0A158NAJ3 A0A2M4AUB9 K7JA85 A0A069DTR7 A0A2R7WPL3 A0A1B0FDY0 A0A1A9WYS5 A0A1Y1KPM3 A0A182GCL4 E9IF49 A0A0R3NN60 A0A0N7ZCF4
PDB
5MBX
E-value=2.19411e-67,
Score=651
Ontologies
GO
Topology
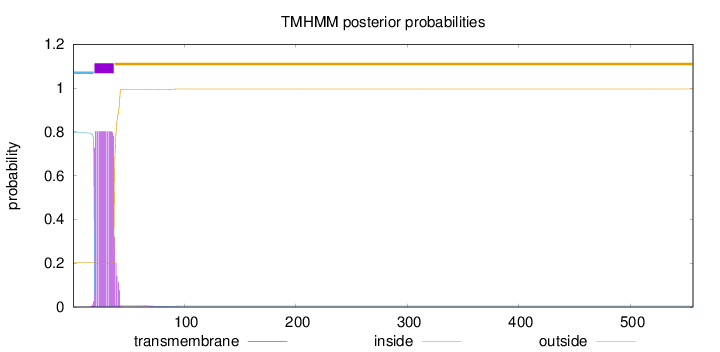
Length:
556
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
15.31556
Exp number, first 60 AAs:
15.24049
Total prob of N-in:
0.79537
POSSIBLE N-term signal
sequence
inside
1 - 19
TMhelix
20 - 37
outside
38 - 556
Population Genetic Test Statistics
Pi
36.711195
Theta
35.37455
Tajima's D
-1.507927
CLR
284.962593
CSRT
0.0584470776461177
Interpretation
Uncertain
