Gene
KWMTBOMO13723 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011504
Annotation
H+_transporting_ATP_synthase_O_subunit_isoform_1_[Bombyx_mori]
Full name
ATP synthase subunit O, mitochondrial
Alternative Name
Oligomycin sensitivity conferral protein
Location in the cell
Mitochondrial Reliability : 2.355
Sequence
CDS
ATGTCGGCTTTAAAGGGGAATCTGCTGGTTCGCTCTTTGAGCACAAGTGTCGCCTCAGCACAAATGGTAAAACCTCCAGTGCAAGTATTTGGATTGGAAGGTCGGTATGCTTCCGCTCTTTTTTCAGCAGCATCAAAGACCAAGGCACTTGACATCGTCGAAAAAGAGCTCGGCCAGTTTCAACAGTCTATCAAAACTGATGCAAAGCTCAAGGAATTCATCATCAACCCGACATTAAAAAGAAGCATGAAGGTAGATGCGTTGAAACATGTCGCCAATAAGATCAGTCTTTCTCCTACAACTGGTAATCTACTTGGATTACTTGCTGAAAATGGTCGCTTGGACAAACTGGAGGCTGTCATTAATGCTTTTAAGATTATGATGGCTGCCCACCGGGGTGAAGTAACTTGTGAAGTTGTTACAGCCAAACCACTAGATCAAGCACAGAGGCAAAATTTGGAAGCAGCGCTTAAGAAATTCCTGAAGGGCAATGAAACTCTGCAGCTTACAGCTAAAGTGGATCCTTCATTGATTGGTGGCATGGTTGTTTCAATTGGGGATAAATATGTCGACATGAGTGTAGCAAGTAAAGTCAAGAAATACACTGAGCTTATCAGTGCTGCTGTTTAA
Protein
MSALKGNLLVRSLSTSVASAQMVKPPVQVFGLEGRYASALFSAASKTKALDIVEKELGQFQQSIKTDAKLKEFIINPTLKRSMKVDALKHVANKISLSPTTGNLLGLLAENGRLDKLEAVINAFKIMMAAHRGEVTCEVVTAKPLDQAQRQNLEAALKKFLKGNETLQLTAKVDPSLIGGMVVSIGDKYVDMSVASKVKKYTELISAAV
Summary
Subunit
F-type ATPases have 2 components, CF(1) - the catalytic core - and CF(0) - the membrane proton channel. CF(1) has five subunits: alpha(3), beta(3), gamma(1), delta(1), epsilon(1). CF(0) has three main subunits: a, b and c.
Similarity
Belongs to the ATPase delta chain family.
Belongs to the nucleosome assembly protein (NAP) family.
Belongs to the nucleosome assembly protein (NAP) family.
Keywords
ATP synthesis
Complete proteome
Hydrogen ion transport
Ion transport
Membrane
Mitochondrion
Mitochondrion inner membrane
Reference proteome
Transit peptide
Transport
Feature
chain ATP synthase subunit O, mitochondrial
Uniprot
Q1HPX3
H9JPP9
Q1HPX2
A0A2A4J874
S4PH25
A0A3G1T1H1
+ More
E7EC49 A0A2H1VHR2 A0A194RPK9 A0A3S2LUU7 A0A212FJK0 I4DKE8 A0A0T6B1X1 A0A0L7LM26 A0A1Y1KN69 D6WA56 A0A084VHA5 A0A023EJM2 U5EXM1 Q7QAP1 A0A182HZC9 A0A182MU44 Q1W2B6 Q0IEE4 A0A2P8YED4 A0A182WVQ9 A0A182VLN5 A0A182TTR0 A0A023EJP6 T1DPQ3 A0A182KXL8 A0A2M3Z0C6 A0A2M4A4H4 A0A2M4BZR3 A0A2M4CTW9 A0A1B6EK86 A0A182P926 A0A2J7QMN5 A0A182JRK0 T1DPR0 A0A182QMJ3 A0A182RPN0 K7J2Q6 A0A1B6K528 A0A182Y7B7 B0XIT5 A0A182VQ73 A0A067RFX8 A0A0L0C9M4 A0A182JK57 A0A182FNS0 R4UJE6 K7J2Q5 A0A232EUS4 E9IFT3 A0A2M4CZ33 W5JJ88 B3MX88 A0A1J1INU8 E1ZY57 D3TMR0 A0A1B0AJ90 B4NIR9 A0A0K8TPA9 A0A3L8DU43 A0A336LUN8 A0A026W2N9 B4JGC5 N6UJY5 A0A182N4Q5 B4PS65 B4HDZ3 B3P0T2 Q24439 A0A1A9X437 T1PFM9 A0A1W4VUW1 A0A1L8E108 A0A0M3QYJ5 A0A0H5B598 A0A0H5AKU0 A0A1W4X734 A0A3B0KFV4 A0A0C9QEN5 A0A034W9S7 A0A195C6D4 A0A1L8EEX5 A0A1I8PDG9 A0A1L8EF97 A0A1L8EF01 W8CB91 A0A1A9X8K5 A0A158NBL9 B4K5V4 A0A1B0D7A7 B4LZZ3 A0A0C9R6B2 Q5XUB9 A0A0K8USI6 A0A195DRA9
E7EC49 A0A2H1VHR2 A0A194RPK9 A0A3S2LUU7 A0A212FJK0 I4DKE8 A0A0T6B1X1 A0A0L7LM26 A0A1Y1KN69 D6WA56 A0A084VHA5 A0A023EJM2 U5EXM1 Q7QAP1 A0A182HZC9 A0A182MU44 Q1W2B6 Q0IEE4 A0A2P8YED4 A0A182WVQ9 A0A182VLN5 A0A182TTR0 A0A023EJP6 T1DPQ3 A0A182KXL8 A0A2M3Z0C6 A0A2M4A4H4 A0A2M4BZR3 A0A2M4CTW9 A0A1B6EK86 A0A182P926 A0A2J7QMN5 A0A182JRK0 T1DPR0 A0A182QMJ3 A0A182RPN0 K7J2Q6 A0A1B6K528 A0A182Y7B7 B0XIT5 A0A182VQ73 A0A067RFX8 A0A0L0C9M4 A0A182JK57 A0A182FNS0 R4UJE6 K7J2Q5 A0A232EUS4 E9IFT3 A0A2M4CZ33 W5JJ88 B3MX88 A0A1J1INU8 E1ZY57 D3TMR0 A0A1B0AJ90 B4NIR9 A0A0K8TPA9 A0A3L8DU43 A0A336LUN8 A0A026W2N9 B4JGC5 N6UJY5 A0A182N4Q5 B4PS65 B4HDZ3 B3P0T2 Q24439 A0A1A9X437 T1PFM9 A0A1W4VUW1 A0A1L8E108 A0A0M3QYJ5 A0A0H5B598 A0A0H5AKU0 A0A1W4X734 A0A3B0KFV4 A0A0C9QEN5 A0A034W9S7 A0A195C6D4 A0A1L8EEX5 A0A1I8PDG9 A0A1L8EF97 A0A1L8EF01 W8CB91 A0A1A9X8K5 A0A158NBL9 B4K5V4 A0A1B0D7A7 B4LZZ3 A0A0C9R6B2 Q5XUB9 A0A0K8USI6 A0A195DRA9
Pubmed
19121390
23622113
26354079
22118469
22651552
26227816
+ More
28004739 18362917 19820115 24438588 24945155 12364791 14747013 17210077 17510324 29403074 26483478 20966253 20075255 25244985 24845553 26108605 28648823 21282665 20920257 23761445 17994087 20798317 20353571 26369729 30249741 24508170 23537049 17550304 10071211 10731132 12537572 12537569 25315136 25348373 24495485 21347285
28004739 18362917 19820115 24438588 24945155 12364791 14747013 17210077 17510324 29403074 26483478 20966253 20075255 25244985 24845553 26108605 28648823 21282665 20920257 23761445 17994087 20798317 20353571 26369729 30249741 24508170 23537049 17550304 10071211 10731132 12537572 12537569 25315136 25348373 24495485 21347285
EMBL
DQ443279
ABF51368.1
BABH01015147
DQ443280
ABF51369.1
NWSH01002449
+ More
PCG68285.1 GAIX01005955 JAA86605.1 MG992387 AXY94825.1 HQ649759 ADV36665.1 ODYU01002594 SOQ40301.1 KQ459833 KPJ19788.1 RSAL01000184 RVE44841.1 AGBW02008253 OWR53927.1 AK401766 KQ459601 BAM18388.1 KPI94045.1 LJIG01016274 KRT81093.1 JTDY01000577 KOB76603.1 GEZM01081401 JAV61650.1 KQ971312 EEZ99200.1 ATLV01013150 KE524842 KFB37349.1 GAPW01004357 JAC09241.1 GANO01002480 JAB57391.1 AAAB01008888 EAA08884.3 APCN01005541 AXCM01000623 DQ445511 GEBQ01032152 ABD98749.1 JAT07825.1 CH477695 EAT37155.1 PYGN01000667 PSN42534.1 JXUM01125166 GAPW01004358 KQ566915 JAC09240.1 KXJ69740.1 GAMD01003144 JAA98446.1 GGFM01001195 MBW21946.1 GGFK01002309 MBW35630.1 GGFJ01009353 MBW58494.1 GGFL01004443 MBW68621.1 GECZ01031467 JAS38302.1 NEVH01013205 PNF29813.1 GAMD01003124 JAA98466.1 AXCN02000015 GECU01001140 JAT06567.1 DS233377 EDS29676.1 KK852498 KDR22637.1 JRES01000819 KNC28149.1 KC632370 AGM32184.1 NNAY01002089 OXU22114.1 GL762897 EFZ20576.1 GGFL01006347 MBW70525.1 ADMH02001019 ETN64422.1 CH902629 EDV35311.1 CVRI01000057 CRL01832.1 GL435171 EFN73906.1 EZ422712 ADD18988.1 CH964272 EDW83783.1 GDAI01001381 JAI16222.1 QOIP01000004 RLU23268.1 UFQT01000197 SSX21570.1 KK107459 EZA50350.1 CH916369 EDV92594.1 APGK01017097 KB739995 KB630277 KB632201 ENN82040.1 ERL83508.1 ERL90057.1 CM000160 EDW97485.1 CH480815 EDW42085.1 CH954181 EDV48980.1 X99666 AE014297 AY058261 KA647464 AFP62093.1 GFDF01001758 JAV12326.1 CP012526 ALC47729.1 LC058926 LC058927 LC058928 LC058929 LC058930 LC058931 LC058932 LC058933 LC058934 LC058935 LC058936 LC058937 LC058938 LC058939 LC058940 LC058941 BAR89771.1 BAR89772.1 BAR89773.1 BAR89774.1 BAR89775.1 BAR89776.1 BAR89777.1 BAR89778.1 BAR89779.1 BAR89780.1 BAR89781.1 BAR89782.1 BAR89783.1 BAR89784.1 BAR89785.1 BAR89786.1 LC058942 LC058943 LC058944 LC058945 LC058946 LC058947 LC058948 LC058949 LC058950 LC058951 LC058952 LC058953 LC058954 LC058955 LC058956 LC058957 BAR89787.1 BAR89788.1 BAR89789.1 BAR89790.1 BAR89791.1 BAR89792.1 BAR89793.1 BAR89794.1 BAR89795.1 BAR89796.1 BAR89797.1 BAR89798.1 BAR89799.1 BAR89800.1 BAR89801.1 BAR89802.1 OUUW01000008 SPP83931.1 GBYB01012968 JAG82735.1 GAKP01007890 JAC51062.1 KQ978219 KYM96407.1 GFDG01001585 JAV17214.1 GFDG01001586 JAV17213.1 GFDG01001584 JAV17215.1 GAMC01005046 JAC01510.1 ADTU01011211 CH933806 EDW15166.1 AJVK01012376 CH940650 EDW67221.1 GBYB01011854 JAG81621.1 AY737535 AAU84928.1 GDHF01022841 JAI29473.1 KQ980581 KYN15347.1
PCG68285.1 GAIX01005955 JAA86605.1 MG992387 AXY94825.1 HQ649759 ADV36665.1 ODYU01002594 SOQ40301.1 KQ459833 KPJ19788.1 RSAL01000184 RVE44841.1 AGBW02008253 OWR53927.1 AK401766 KQ459601 BAM18388.1 KPI94045.1 LJIG01016274 KRT81093.1 JTDY01000577 KOB76603.1 GEZM01081401 JAV61650.1 KQ971312 EEZ99200.1 ATLV01013150 KE524842 KFB37349.1 GAPW01004357 JAC09241.1 GANO01002480 JAB57391.1 AAAB01008888 EAA08884.3 APCN01005541 AXCM01000623 DQ445511 GEBQ01032152 ABD98749.1 JAT07825.1 CH477695 EAT37155.1 PYGN01000667 PSN42534.1 JXUM01125166 GAPW01004358 KQ566915 JAC09240.1 KXJ69740.1 GAMD01003144 JAA98446.1 GGFM01001195 MBW21946.1 GGFK01002309 MBW35630.1 GGFJ01009353 MBW58494.1 GGFL01004443 MBW68621.1 GECZ01031467 JAS38302.1 NEVH01013205 PNF29813.1 GAMD01003124 JAA98466.1 AXCN02000015 GECU01001140 JAT06567.1 DS233377 EDS29676.1 KK852498 KDR22637.1 JRES01000819 KNC28149.1 KC632370 AGM32184.1 NNAY01002089 OXU22114.1 GL762897 EFZ20576.1 GGFL01006347 MBW70525.1 ADMH02001019 ETN64422.1 CH902629 EDV35311.1 CVRI01000057 CRL01832.1 GL435171 EFN73906.1 EZ422712 ADD18988.1 CH964272 EDW83783.1 GDAI01001381 JAI16222.1 QOIP01000004 RLU23268.1 UFQT01000197 SSX21570.1 KK107459 EZA50350.1 CH916369 EDV92594.1 APGK01017097 KB739995 KB630277 KB632201 ENN82040.1 ERL83508.1 ERL90057.1 CM000160 EDW97485.1 CH480815 EDW42085.1 CH954181 EDV48980.1 X99666 AE014297 AY058261 KA647464 AFP62093.1 GFDF01001758 JAV12326.1 CP012526 ALC47729.1 LC058926 LC058927 LC058928 LC058929 LC058930 LC058931 LC058932 LC058933 LC058934 LC058935 LC058936 LC058937 LC058938 LC058939 LC058940 LC058941 BAR89771.1 BAR89772.1 BAR89773.1 BAR89774.1 BAR89775.1 BAR89776.1 BAR89777.1 BAR89778.1 BAR89779.1 BAR89780.1 BAR89781.1 BAR89782.1 BAR89783.1 BAR89784.1 BAR89785.1 BAR89786.1 LC058942 LC058943 LC058944 LC058945 LC058946 LC058947 LC058948 LC058949 LC058950 LC058951 LC058952 LC058953 LC058954 LC058955 LC058956 LC058957 BAR89787.1 BAR89788.1 BAR89789.1 BAR89790.1 BAR89791.1 BAR89792.1 BAR89793.1 BAR89794.1 BAR89795.1 BAR89796.1 BAR89797.1 BAR89798.1 BAR89799.1 BAR89800.1 BAR89801.1 BAR89802.1 OUUW01000008 SPP83931.1 GBYB01012968 JAG82735.1 GAKP01007890 JAC51062.1 KQ978219 KYM96407.1 GFDG01001585 JAV17214.1 GFDG01001586 JAV17213.1 GFDG01001584 JAV17215.1 GAMC01005046 JAC01510.1 ADTU01011211 CH933806 EDW15166.1 AJVK01012376 CH940650 EDW67221.1 GBYB01011854 JAG81621.1 AY737535 AAU84928.1 GDHF01022841 JAI29473.1 KQ980581 KYN15347.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000283053
UP000007151
UP000053268
+ More
UP000037510 UP000007266 UP000030765 UP000007062 UP000075840 UP000075883 UP000008820 UP000245037 UP000076407 UP000075903 UP000075902 UP000069940 UP000249989 UP000075882 UP000075885 UP000235965 UP000075881 UP000075886 UP000075900 UP000002358 UP000076408 UP000002320 UP000075920 UP000027135 UP000037069 UP000075880 UP000069272 UP000215335 UP000000673 UP000007801 UP000183832 UP000000311 UP000092445 UP000007798 UP000279307 UP000053097 UP000001070 UP000019118 UP000030742 UP000075884 UP000002282 UP000001292 UP000008711 UP000000803 UP000091820 UP000095301 UP000192221 UP000092553 UP000192223 UP000268350 UP000078542 UP000095300 UP000092443 UP000005205 UP000009192 UP000092462 UP000008792 UP000078492
UP000037510 UP000007266 UP000030765 UP000007062 UP000075840 UP000075883 UP000008820 UP000245037 UP000076407 UP000075903 UP000075902 UP000069940 UP000249989 UP000075882 UP000075885 UP000235965 UP000075881 UP000075886 UP000075900 UP000002358 UP000076408 UP000002320 UP000075920 UP000027135 UP000037069 UP000075880 UP000069272 UP000215335 UP000000673 UP000007801 UP000183832 UP000000311 UP000092445 UP000007798 UP000279307 UP000053097 UP000001070 UP000019118 UP000030742 UP000075884 UP000002282 UP000001292 UP000008711 UP000000803 UP000091820 UP000095301 UP000192221 UP000092553 UP000192223 UP000268350 UP000078542 UP000095300 UP000092443 UP000005205 UP000009192 UP000092462 UP000008792 UP000078492
Interpro
Gene 3D
CDD
ProteinModelPortal
Q1HPX3
H9JPP9
Q1HPX2
A0A2A4J874
S4PH25
A0A3G1T1H1
+ More
E7EC49 A0A2H1VHR2 A0A194RPK9 A0A3S2LUU7 A0A212FJK0 I4DKE8 A0A0T6B1X1 A0A0L7LM26 A0A1Y1KN69 D6WA56 A0A084VHA5 A0A023EJM2 U5EXM1 Q7QAP1 A0A182HZC9 A0A182MU44 Q1W2B6 Q0IEE4 A0A2P8YED4 A0A182WVQ9 A0A182VLN5 A0A182TTR0 A0A023EJP6 T1DPQ3 A0A182KXL8 A0A2M3Z0C6 A0A2M4A4H4 A0A2M4BZR3 A0A2M4CTW9 A0A1B6EK86 A0A182P926 A0A2J7QMN5 A0A182JRK0 T1DPR0 A0A182QMJ3 A0A182RPN0 K7J2Q6 A0A1B6K528 A0A182Y7B7 B0XIT5 A0A182VQ73 A0A067RFX8 A0A0L0C9M4 A0A182JK57 A0A182FNS0 R4UJE6 K7J2Q5 A0A232EUS4 E9IFT3 A0A2M4CZ33 W5JJ88 B3MX88 A0A1J1INU8 E1ZY57 D3TMR0 A0A1B0AJ90 B4NIR9 A0A0K8TPA9 A0A3L8DU43 A0A336LUN8 A0A026W2N9 B4JGC5 N6UJY5 A0A182N4Q5 B4PS65 B4HDZ3 B3P0T2 Q24439 A0A1A9X437 T1PFM9 A0A1W4VUW1 A0A1L8E108 A0A0M3QYJ5 A0A0H5B598 A0A0H5AKU0 A0A1W4X734 A0A3B0KFV4 A0A0C9QEN5 A0A034W9S7 A0A195C6D4 A0A1L8EEX5 A0A1I8PDG9 A0A1L8EF97 A0A1L8EF01 W8CB91 A0A1A9X8K5 A0A158NBL9 B4K5V4 A0A1B0D7A7 B4LZZ3 A0A0C9R6B2 Q5XUB9 A0A0K8USI6 A0A195DRA9
E7EC49 A0A2H1VHR2 A0A194RPK9 A0A3S2LUU7 A0A212FJK0 I4DKE8 A0A0T6B1X1 A0A0L7LM26 A0A1Y1KN69 D6WA56 A0A084VHA5 A0A023EJM2 U5EXM1 Q7QAP1 A0A182HZC9 A0A182MU44 Q1W2B6 Q0IEE4 A0A2P8YED4 A0A182WVQ9 A0A182VLN5 A0A182TTR0 A0A023EJP6 T1DPQ3 A0A182KXL8 A0A2M3Z0C6 A0A2M4A4H4 A0A2M4BZR3 A0A2M4CTW9 A0A1B6EK86 A0A182P926 A0A2J7QMN5 A0A182JRK0 T1DPR0 A0A182QMJ3 A0A182RPN0 K7J2Q6 A0A1B6K528 A0A182Y7B7 B0XIT5 A0A182VQ73 A0A067RFX8 A0A0L0C9M4 A0A182JK57 A0A182FNS0 R4UJE6 K7J2Q5 A0A232EUS4 E9IFT3 A0A2M4CZ33 W5JJ88 B3MX88 A0A1J1INU8 E1ZY57 D3TMR0 A0A1B0AJ90 B4NIR9 A0A0K8TPA9 A0A3L8DU43 A0A336LUN8 A0A026W2N9 B4JGC5 N6UJY5 A0A182N4Q5 B4PS65 B4HDZ3 B3P0T2 Q24439 A0A1A9X437 T1PFM9 A0A1W4VUW1 A0A1L8E108 A0A0M3QYJ5 A0A0H5B598 A0A0H5AKU0 A0A1W4X734 A0A3B0KFV4 A0A0C9QEN5 A0A034W9S7 A0A195C6D4 A0A1L8EEX5 A0A1I8PDG9 A0A1L8EF97 A0A1L8EF01 W8CB91 A0A1A9X8K5 A0A158NBL9 B4K5V4 A0A1B0D7A7 B4LZZ3 A0A0C9R6B2 Q5XUB9 A0A0K8USI6 A0A195DRA9
PDB
5FIL
E-value=1.67895e-41,
Score=422
Ontologies
PATHWAY
GO
Topology
Subcellular location
Mitochondrion
Mitochondrion inner membrane
Mitochondrion inner membrane
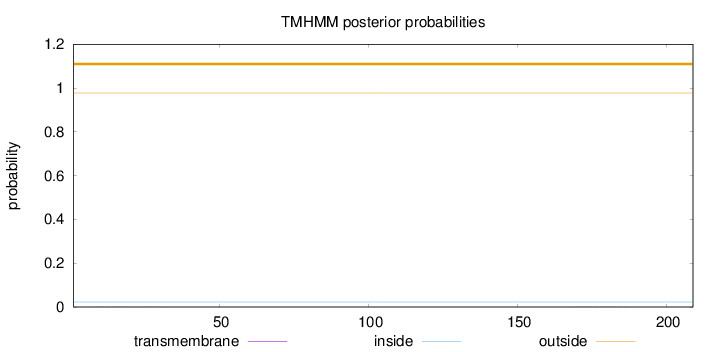
Length:
209
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01174
Exp number, first 60 AAs:
0.00672
Total prob of N-in:
0.02293
outside
1 - 209
Population Genetic Test Statistics
Pi
27.704472
Theta
180.799577
Tajima's D
1.936578
CLR
0.401496
CSRT
0.872406379681016
Interpretation
Uncertain
