Gene
KWMTBOMO13721 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011508
Annotation
chaperonin_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 3.406
Sequence
CDS
ATGGCTCTACATGTACCGAAAGCTCCAGGAGTACCTCAGATGCTAAAAGATGGTGCCCGGATGTTTTCTGGATTGGAAGAGGCAGTATATAGAAATATTAATGCCTGTAAACAATTTGCACAGAGTGTGCGTTCTGCATATGGCCCTAATGGCATGAACAAAATGATCATCAATCATATTGATAAGCAATTTGTTACCAGTGATGCTGGCACCATCATAAGAGAGTTAGATGTGGAACATCCAGCAGCCAAACTAATGGTACTAGCTAGTCAAATGCAAGATGCTGAAGTTGGTGATGGTACCAACTTTGTGATTGTCTTGTCAGGAGCATTATTAGAAGCTGCAGAGGAACTACTCCGCTTGGGAGTAACTACCAGTGAAATTGCTGATGGATATGAGAGAGCACTTGACAAATGTCTGGAAATCCTTCCTTCTCTCATTTGTAATGAGATAAAAGACACAAAAAACATTAAAGAAGTTACAAAGGGTATTGTTGCTGCTATCATGTCTAAACAATATGGTCATGAACAATTCATTTCTGAGCTTGTCACGAAAGCCTGTGTTGCAATACTACCAGAAAAAACTACATTTAATGTTGATAATGTTAGAGTTTGCAAAATTCTAGGTGCTGGACTGTTGCAATCTGAGGTACTTTCTGGAATGGTATTTAGACGGGAAGTAGAAGGAGATGTCTCTTCAGCTAAGAATGCTAAAATAGCAGTATACTCATGTCCTATTGACATAATCCAGACTGAAACTAAGGGCACGGTCCTCATTAAGAGTGCTGATGAATTACTGAACTTCAGTAAAGGAGAAGAATCTTTACTTGAAAAGCAGATTAAAGATATTTCCGACTCTGGGGTGAAAGTTATAGTGGCTGGTGCTAAATTTGGAGATATGGCCTTACACTTCTTGAATAAATACAATATTATGGCTGTCCGTCTTAATTCCAAATTCGACATTCGTCGGTTGGCTAAGACTGTAAATGCTACGGTTTTACCAAGATTAACGACCCCAACTACCCAGGAACTTGGATACTGTGATGCTGTACATGTTGATGAAGTCGGGGACACTCGTGTTGTTGTGTTCAAGATGGAAAGCTCAGAATCTCGGATTTCCACTATTGTCATTAGAGGTTCTACTGACAACTACATGGACGATATAGAGAGGGCTATTGATGATGGTGTTAATACATTCAAGGGAATTGCTAGAGATGGGAGATTTGTACCAGGCGCCGGAGCAACAGAAATTGAATTAGCACAACAGTTATTGGAGTTTGCTGACACTCTGCCCGGTTTAGAACAGTATGCTGTAAGGAAGTTTGCAGTGGCTCTTGAGAGCATACCTCATGCTTTGGCAGATAACTCGGGTGCCAATGCCACTGAAGTTGTAAACAAAATTTACAAGGCACACAAGGAGGGACAAAAAAATGCTGGTTTTGATATTGATTCAGAAAACAATGGTGTTTGTGATGCAAAAGAAAAAGGCATAATTGACTCATATCTTTTGAAATTCTGGGGAATAAAGTATGCTGTTGGAGCAGCAACAACCATTCTTAAGGTTGATCAAATCATTATGGCAAAGAGAGCAGGCGGACCAAAACCAAGAGCGCCAGCTGGAAGTGATGATGAGTCTTAA
Protein
MALHVPKAPGVPQMLKDGARMFSGLEEAVYRNINACKQFAQSVRSAYGPNGMNKMIINHIDKQFVTSDAGTIIRELDVEHPAAKLMVLASQMQDAEVGDGTNFVIVLSGALLEAAEELLRLGVTTSEIADGYERALDKCLEILPSLICNEIKDTKNIKEVTKGIVAAIMSKQYGHEQFISELVTKACVAILPEKTTFNVDNVRVCKILGAGLLQSEVLSGMVFRREVEGDVSSAKNAKIAVYSCPIDIIQTETKGTVLIKSADELLNFSKGEESLLEKQIKDISDSGVKVIVAGAKFGDMALHFLNKYNIMAVRLNSKFDIRRLAKTVNATVLPRLTTPTTQELGYCDAVHVDEVGDTRVVVFKMESSESRISTIVIRGSTDNYMDDIERAIDDGVNTFKGIARDGRFVPGAGATEIELAQQLLEFADTLPGLEQYAVRKFAVALESIPHALADNSGANATEVVNKIYKAHKEGQKNAGFDIDSENNGVCDAKEKGIIDSYLLKFWGIKYAVGAATTILKVDQIIMAKRAGGPKPRAPAGSDDES
Summary
Similarity
Belongs to the TCP-1 chaperonin family.
Uniprot
A1YM11
A0A2H1VHH7
A0A2A4J936
A0A212FJL1
A0A2U9Q1E0
A0A3S2LEB1
+ More
S4P9I3 A0A194RUH2 I4DJL3 A0A0L7LLG0 D6W9Y2 A0A023EUU8 A0A1Q3F2E7 B0WG55 A0A023ETT8 V5GVF8 A0A1S4FXE3 U5EWC2 Q16L72 A0A182MYE6 A0A182Q5N4 A0A034VX78 A0A0A1X516 A0A1Y1KZD3 T1PEA6 A0A1W4X519 A0A084WI50 W8BKD8 A0A182JJ62 A0A0K8W3C1 A0A2M3Z348 A0A1B6DPY0 J3JVY6 B4GCT8 A0A182WCV1 A0A182V7S8 A0A182X1T8 A0A182LQ38 Q7Q8S5 A0A182HFQ3 A0A182MW10 A0A182YA44 A0A182K4Y4 A0A182P8G2 Q28Y76 A0A0L0C5U2 A0A2M4BJC5 A0A182UH82 A0A2M4A8U1 A0A1W4UXS3 A0A182RLP0 B4KTQ8 A0A1J1HLW7 W5JAC0 A0A182FV10 T1DFU4 A0A0K8TSQ8 A0A1L8ED20 A0A1I8PPQ7 A0A1L8ECV3 A0A336MKM4 A0A0M4E3Z2 B4MQW0 A0A1B0GLZ2 B4LNN4 A0A1L8DUS8 A0A1L8DU97 B3MFD3 V5RE76 B4JVE5 A0A1B0BUD0 A0A232EYC7 K7INB6 A0A3B0JRZ5 B4QGA3 F4WF07 B3N850 A0A151WXD5 A0A195AYK2 A0A195DYB9 B4P3D7 B4HSA8 E9IM02 A0A158NP98 N6UUA0 A0A026W243 A0A0K8W754 Q7K3J0 E0VHJ5 A0A1B6J8I8 A0A0L7QQP1 A0A2A3EAD8 A0A1B6L1Y9 A0A088A892 A0A0M9A3K7 A0A067RGA7 A0A1A9XIC8 A0A0C9RXL2 A0A2J7RBZ0 A0A1A9WQX4
S4P9I3 A0A194RUH2 I4DJL3 A0A0L7LLG0 D6W9Y2 A0A023EUU8 A0A1Q3F2E7 B0WG55 A0A023ETT8 V5GVF8 A0A1S4FXE3 U5EWC2 Q16L72 A0A182MYE6 A0A182Q5N4 A0A034VX78 A0A0A1X516 A0A1Y1KZD3 T1PEA6 A0A1W4X519 A0A084WI50 W8BKD8 A0A182JJ62 A0A0K8W3C1 A0A2M3Z348 A0A1B6DPY0 J3JVY6 B4GCT8 A0A182WCV1 A0A182V7S8 A0A182X1T8 A0A182LQ38 Q7Q8S5 A0A182HFQ3 A0A182MW10 A0A182YA44 A0A182K4Y4 A0A182P8G2 Q28Y76 A0A0L0C5U2 A0A2M4BJC5 A0A182UH82 A0A2M4A8U1 A0A1W4UXS3 A0A182RLP0 B4KTQ8 A0A1J1HLW7 W5JAC0 A0A182FV10 T1DFU4 A0A0K8TSQ8 A0A1L8ED20 A0A1I8PPQ7 A0A1L8ECV3 A0A336MKM4 A0A0M4E3Z2 B4MQW0 A0A1B0GLZ2 B4LNN4 A0A1L8DUS8 A0A1L8DU97 B3MFD3 V5RE76 B4JVE5 A0A1B0BUD0 A0A232EYC7 K7INB6 A0A3B0JRZ5 B4QGA3 F4WF07 B3N850 A0A151WXD5 A0A195AYK2 A0A195DYB9 B4P3D7 B4HSA8 E9IM02 A0A158NP98 N6UUA0 A0A026W243 A0A0K8W754 Q7K3J0 E0VHJ5 A0A1B6J8I8 A0A0L7QQP1 A0A2A3EAD8 A0A1B6L1Y9 A0A088A892 A0A0M9A3K7 A0A067RGA7 A0A1A9XIC8 A0A0C9RXL2 A0A2J7RBZ0 A0A1A9WQX4
Pubmed
22118469
23622113
26354079
22651552
26227816
18362917
+ More
19820115 24945155 17510324 25348373 25830018 28004739 25315136 24438588 24495485 22516182 17994087 20966253 12364791 14747013 17210077 25244985 15632085 26108605 20920257 23761445 26369729 28648823 20075255 22936249 21719571 17550304 21282665 21347285 23537049 24508170 30249741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20566863 24845553
19820115 24945155 17510324 25348373 25830018 28004739 25315136 24438588 24495485 22516182 17994087 20966253 12364791 14747013 17210077 25244985 15632085 26108605 20920257 23761445 26369729 28648823 20075255 22936249 21719571 17550304 21282665 21347285 23537049 24508170 30249741 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20566863 24845553
EMBL
EF105369
ABM05494.1
ODYU01002594
SOQ40298.1
NWSH01002449
PCG68276.1
+ More
AGBW02008253 OWR53925.1 MF471349 MF471350 AWT57841.1 AWT57842.1 RSAL01000184 RVE44843.1 GAIX01003744 JAA88816.1 KQ459833 KPJ19786.1 AK401481 KQ459601 BAM18103.1 KPI94047.1 JTDY01000698 KOB76179.1 KQ971312 EEZ98562.1 GAPW01000836 JAC12762.1 GFDL01013323 JAV21722.1 DS231923 EDS26751.1 GAPW01000835 JAC12763.1 GALX01004248 JAB64218.1 GANO01001487 JAB58384.1 CH477919 EAT35055.1 AXCN02001048 GAKP01011903 JAC47049.1 GBXI01007878 JAD06414.1 GEZM01068805 JAV66762.1 KA647071 AFP61700.1 ATLV01023913 KE525347 KFB49894.1 GAMC01016526 JAB90029.1 GDHF01006636 JAI45678.1 GGFM01002117 MBW22868.1 GEDC01009579 JAS27719.1 BT127404 AEE62366.1 CH479181 EDW32501.1 AAAB01008933 EAA09922.3 APCN01005563 AXCM01000498 CM000071 EAL26089.1 JRES01000848 KNC27763.1 GGFJ01004019 MBW53160.1 GGFK01003902 MBW37223.1 CH933808 EDW09641.1 CVRI01000008 CRK88530.1 ADMH02002040 ETN59805.1 GAMD01003026 JAA98564.1 GDAI01000440 JAI17163.1 GFDG01002253 JAV16546.1 GFDG01002252 JAV16547.1 UFQS01001192 UFQT01001192 SSX09617.1 SSX29413.1 CP012524 ALC40641.1 CH963849 EDW74499.1 AJVK01009183 AJVK01009184 CH940648 EDW60104.1 GFDF01004059 JAV10025.1 GFDF01004060 JAV10024.1 CH902619 EDV35607.1 KF621130 AHB33466.1 CH916375 EDV98413.1 JXJN01020612 JXJN01022120 NNAY01001642 OXU23343.1 OUUW01000001 SPP73888.1 CM000362 CM002911 EDX06253.1 KMY92366.1 GL888115 EGI67058.1 CH954177 EDV59463.1 KQ982668 KYQ52508.1 KQ976703 KYM77120.1 KQ980066 KYN17905.1 CM000157 EDW89410.1 CH480816 EDW47003.1 GL764129 EFZ18367.1 ADTU01022285 ADTU01022286 APGK01013773 APGK01035524 KB740927 KB739110 ENN77994.1 ENN82382.1 KK107474 QOIP01000002 EZA50150.1 RLU25833.1 GDHF01005614 JAI46700.1 AE013599 AY058545 AAF59029.3 AAL13774.1 DS235171 EEB12881.1 GECU01012209 JAS95497.1 KQ414786 KOC60942.1 KZ288325 PBC28031.1 GEBQ01022260 GEBQ01017988 JAT17717.1 JAT21989.1 KQ435747 KOX76507.1 KK852482 KDR22911.1 GBYB01013915 JAG83682.1 NEVH01005889 PNF38356.1
AGBW02008253 OWR53925.1 MF471349 MF471350 AWT57841.1 AWT57842.1 RSAL01000184 RVE44843.1 GAIX01003744 JAA88816.1 KQ459833 KPJ19786.1 AK401481 KQ459601 BAM18103.1 KPI94047.1 JTDY01000698 KOB76179.1 KQ971312 EEZ98562.1 GAPW01000836 JAC12762.1 GFDL01013323 JAV21722.1 DS231923 EDS26751.1 GAPW01000835 JAC12763.1 GALX01004248 JAB64218.1 GANO01001487 JAB58384.1 CH477919 EAT35055.1 AXCN02001048 GAKP01011903 JAC47049.1 GBXI01007878 JAD06414.1 GEZM01068805 JAV66762.1 KA647071 AFP61700.1 ATLV01023913 KE525347 KFB49894.1 GAMC01016526 JAB90029.1 GDHF01006636 JAI45678.1 GGFM01002117 MBW22868.1 GEDC01009579 JAS27719.1 BT127404 AEE62366.1 CH479181 EDW32501.1 AAAB01008933 EAA09922.3 APCN01005563 AXCM01000498 CM000071 EAL26089.1 JRES01000848 KNC27763.1 GGFJ01004019 MBW53160.1 GGFK01003902 MBW37223.1 CH933808 EDW09641.1 CVRI01000008 CRK88530.1 ADMH02002040 ETN59805.1 GAMD01003026 JAA98564.1 GDAI01000440 JAI17163.1 GFDG01002253 JAV16546.1 GFDG01002252 JAV16547.1 UFQS01001192 UFQT01001192 SSX09617.1 SSX29413.1 CP012524 ALC40641.1 CH963849 EDW74499.1 AJVK01009183 AJVK01009184 CH940648 EDW60104.1 GFDF01004059 JAV10025.1 GFDF01004060 JAV10024.1 CH902619 EDV35607.1 KF621130 AHB33466.1 CH916375 EDV98413.1 JXJN01020612 JXJN01022120 NNAY01001642 OXU23343.1 OUUW01000001 SPP73888.1 CM000362 CM002911 EDX06253.1 KMY92366.1 GL888115 EGI67058.1 CH954177 EDV59463.1 KQ982668 KYQ52508.1 KQ976703 KYM77120.1 KQ980066 KYN17905.1 CM000157 EDW89410.1 CH480816 EDW47003.1 GL764129 EFZ18367.1 ADTU01022285 ADTU01022286 APGK01013773 APGK01035524 KB740927 KB739110 ENN77994.1 ENN82382.1 KK107474 QOIP01000002 EZA50150.1 RLU25833.1 GDHF01005614 JAI46700.1 AE013599 AY058545 AAF59029.3 AAL13774.1 DS235171 EEB12881.1 GECU01012209 JAS95497.1 KQ414786 KOC60942.1 KZ288325 PBC28031.1 GEBQ01022260 GEBQ01017988 JAT17717.1 JAT21989.1 KQ435747 KOX76507.1 KK852482 KDR22911.1 GBYB01013915 JAG83682.1 NEVH01005889 PNF38356.1
Proteomes
UP000218220
UP000007151
UP000283053
UP000053240
UP000053268
UP000037510
+ More
UP000007266 UP000002320 UP000008820 UP000075884 UP000075886 UP000095301 UP000192223 UP000030765 UP000075880 UP000008744 UP000075920 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000075883 UP000076408 UP000075881 UP000075885 UP000001819 UP000037069 UP000075902 UP000192221 UP000075900 UP000009192 UP000183832 UP000000673 UP000069272 UP000095300 UP000092553 UP000007798 UP000092462 UP000008792 UP000007801 UP000001070 UP000092460 UP000215335 UP000002358 UP000268350 UP000000304 UP000007755 UP000008711 UP000075809 UP000078540 UP000078492 UP000002282 UP000001292 UP000005205 UP000019118 UP000053097 UP000279307 UP000000803 UP000009046 UP000053825 UP000242457 UP000005203 UP000053105 UP000027135 UP000092443 UP000235965 UP000091820
UP000007266 UP000002320 UP000008820 UP000075884 UP000075886 UP000095301 UP000192223 UP000030765 UP000075880 UP000008744 UP000075920 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000075883 UP000076408 UP000075881 UP000075885 UP000001819 UP000037069 UP000075902 UP000192221 UP000075900 UP000009192 UP000183832 UP000000673 UP000069272 UP000095300 UP000092553 UP000007798 UP000092462 UP000008792 UP000007801 UP000001070 UP000092460 UP000215335 UP000002358 UP000268350 UP000000304 UP000007755 UP000008711 UP000075809 UP000078540 UP000078492 UP000002282 UP000001292 UP000005205 UP000019118 UP000053097 UP000279307 UP000000803 UP000009046 UP000053825 UP000242457 UP000005203 UP000053105 UP000027135 UP000092443 UP000235965 UP000091820
PRIDE
Interpro
IPR002194
Chaperonin_TCP-1_CS
+ More
IPR012721 Chap_CCT_theta
IPR027413 GROEL-like_equatorial_sf
IPR027409 GroEL-like_apical_dom_sf
IPR002423 Cpn60/TCP-1
IPR027410 TCP-1-like_intermed_sf
IPR017998 Chaperone_TCP-1
IPR001969 Aspartic_peptidase_AS
IPR016082 Ribosomal_L30_ferredoxin-like
IPR036919 L30_ferredoxin-like_sf
IPR012721 Chap_CCT_theta
IPR027413 GROEL-like_equatorial_sf
IPR027409 GroEL-like_apical_dom_sf
IPR002423 Cpn60/TCP-1
IPR027410 TCP-1-like_intermed_sf
IPR017998 Chaperone_TCP-1
IPR001969 Aspartic_peptidase_AS
IPR016082 Ribosomal_L30_ferredoxin-like
IPR036919 L30_ferredoxin-like_sf
Gene 3D
ProteinModelPortal
A1YM11
A0A2H1VHH7
A0A2A4J936
A0A212FJL1
A0A2U9Q1E0
A0A3S2LEB1
+ More
S4P9I3 A0A194RUH2 I4DJL3 A0A0L7LLG0 D6W9Y2 A0A023EUU8 A0A1Q3F2E7 B0WG55 A0A023ETT8 V5GVF8 A0A1S4FXE3 U5EWC2 Q16L72 A0A182MYE6 A0A182Q5N4 A0A034VX78 A0A0A1X516 A0A1Y1KZD3 T1PEA6 A0A1W4X519 A0A084WI50 W8BKD8 A0A182JJ62 A0A0K8W3C1 A0A2M3Z348 A0A1B6DPY0 J3JVY6 B4GCT8 A0A182WCV1 A0A182V7S8 A0A182X1T8 A0A182LQ38 Q7Q8S5 A0A182HFQ3 A0A182MW10 A0A182YA44 A0A182K4Y4 A0A182P8G2 Q28Y76 A0A0L0C5U2 A0A2M4BJC5 A0A182UH82 A0A2M4A8U1 A0A1W4UXS3 A0A182RLP0 B4KTQ8 A0A1J1HLW7 W5JAC0 A0A182FV10 T1DFU4 A0A0K8TSQ8 A0A1L8ED20 A0A1I8PPQ7 A0A1L8ECV3 A0A336MKM4 A0A0M4E3Z2 B4MQW0 A0A1B0GLZ2 B4LNN4 A0A1L8DUS8 A0A1L8DU97 B3MFD3 V5RE76 B4JVE5 A0A1B0BUD0 A0A232EYC7 K7INB6 A0A3B0JRZ5 B4QGA3 F4WF07 B3N850 A0A151WXD5 A0A195AYK2 A0A195DYB9 B4P3D7 B4HSA8 E9IM02 A0A158NP98 N6UUA0 A0A026W243 A0A0K8W754 Q7K3J0 E0VHJ5 A0A1B6J8I8 A0A0L7QQP1 A0A2A3EAD8 A0A1B6L1Y9 A0A088A892 A0A0M9A3K7 A0A067RGA7 A0A1A9XIC8 A0A0C9RXL2 A0A2J7RBZ0 A0A1A9WQX4
S4P9I3 A0A194RUH2 I4DJL3 A0A0L7LLG0 D6W9Y2 A0A023EUU8 A0A1Q3F2E7 B0WG55 A0A023ETT8 V5GVF8 A0A1S4FXE3 U5EWC2 Q16L72 A0A182MYE6 A0A182Q5N4 A0A034VX78 A0A0A1X516 A0A1Y1KZD3 T1PEA6 A0A1W4X519 A0A084WI50 W8BKD8 A0A182JJ62 A0A0K8W3C1 A0A2M3Z348 A0A1B6DPY0 J3JVY6 B4GCT8 A0A182WCV1 A0A182V7S8 A0A182X1T8 A0A182LQ38 Q7Q8S5 A0A182HFQ3 A0A182MW10 A0A182YA44 A0A182K4Y4 A0A182P8G2 Q28Y76 A0A0L0C5U2 A0A2M4BJC5 A0A182UH82 A0A2M4A8U1 A0A1W4UXS3 A0A182RLP0 B4KTQ8 A0A1J1HLW7 W5JAC0 A0A182FV10 T1DFU4 A0A0K8TSQ8 A0A1L8ED20 A0A1I8PPQ7 A0A1L8ECV3 A0A336MKM4 A0A0M4E3Z2 B4MQW0 A0A1B0GLZ2 B4LNN4 A0A1L8DUS8 A0A1L8DU97 B3MFD3 V5RE76 B4JVE5 A0A1B0BUD0 A0A232EYC7 K7INB6 A0A3B0JRZ5 B4QGA3 F4WF07 B3N850 A0A151WXD5 A0A195AYK2 A0A195DYB9 B4P3D7 B4HSA8 E9IM02 A0A158NP98 N6UUA0 A0A026W243 A0A0K8W754 Q7K3J0 E0VHJ5 A0A1B6J8I8 A0A0L7QQP1 A0A2A3EAD8 A0A1B6L1Y9 A0A088A892 A0A0M9A3K7 A0A067RGA7 A0A1A9XIC8 A0A0C9RXL2 A0A2J7RBZ0 A0A1A9WQX4
PDB
4B2T
E-value=0,
Score=1771
Ontologies
GO
Topology
Subcellular location
Cytoplasm
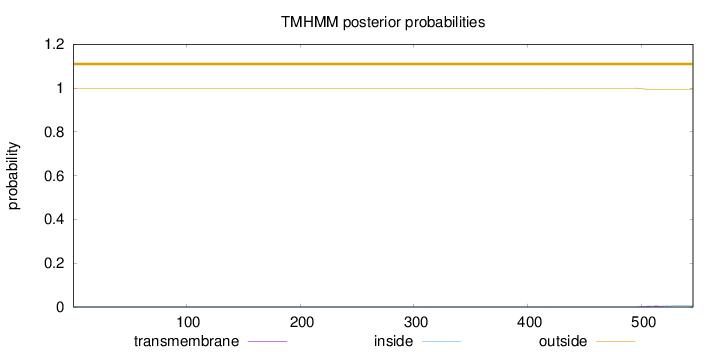
Length:
545
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.12509
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00049
outside
1 - 545
Population Genetic Test Statistics
Pi
3.226174
Theta
32.870942
Tajima's D
-1.074347
CLR
257.127993
CSRT
0.124793760311984
Interpretation
Uncertain
