Gene
KWMTBOMO13717
Pre Gene Modal
BGIBMGA014053
Annotation
PREDICTED:_pancreatic_lipase-related_protein_2-like_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.189 PlasmaMembrane Reliability : 1.652
Sequence
CDS
ATGTGCCGACATAGGGTAATCGTGCTGATAGCTGTTCACTTTATGGTCATGGCACAAGGTGCCACCCTGAATACAGTGCGCCGTTCAGAACCTGAGCTAAGCTTCGTGGAGAACATTTACGTCAGATTATCCGGTGCATGTAGAGCAATCATGGATTTAAGCCATAGCGTTTCGCTAACACAAGTAGACATGATAAGGACGTTAAAAATAATCCAATTAGATGGAAGCACAATGAGAACTTTTCCTATTAATGAAGCATTTGTCAATTTAACAAAACCGGCCATGTTCGATATCACGAAACAAGTTAAAATTATTATTCATGGTTACAAAGACAGTTCACAATCGGCTGTTCCCTTGGAACTTGCAAAATCGTACAATTCGAAAGGGATGTTTAACGTATTTTTGGTCGATGCAGAAGACATGATGAATAAACGATACATTACTTGCGTTCACAATGCTCGTCTCATAGGTAAAAGATTAGCTAATTTATTGGCTAATTTAGAGAATTTTGGGGCGAATGCTGATGATTTCCATCTTTTGGGAATTAGTCTAGGTGCACACATAGCTGGTTGGGCGGGTAAATATTTCCGACTGTATAAACGGCGCTCTATAGGACGCATAACTGGACTAGATCCTGCAGGGCCATGTTTTTCACATGCTTATAGTGATCAAAGATTAGATAAAAGTGATGCTCTTTATGTTGATGTTTTGCATTCTAATAGATTAGTTCAAGGAATCATTGAACCTCTTGGACACGCAGACTTTTATATTAACGGTGGTGGACCTAATCAACCCGGTTGTATGATGCCATCCTGCAGTCATTTACGTGCAGCCCAAGTCTATACAGAGAGTGTTACCACACCTAAATCTTTTGTGGGTATCAGATGTCAAAGTTGGAAGCATTTTGAAGCAAATGCCTGTGAAAAAAACATGTATGCTGTCCTAGGATATGGTTCTTCAACAACATCGAGAGGTTTGTACTACCTAAGAACTTCTGGAAGTCCTCCATTCGGTCTCGGAATGGAAGGAACCAAAATTGTAGGTCCTACATTTGAAAGTTGGTTACGGACTTTGAATATAGCATAA
Protein
MCRHRVIVLIAVHFMVMAQGATLNTVRRSEPELSFVENIYVRLSGACRAIMDLSHSVSLTQVDMIRTLKIIQLDGSTMRTFPINEAFVNLTKPAMFDITKQVKIIIHGYKDSSQSAVPLELAKSYNSKGMFNVFLVDAEDMMNKRYITCVHNARLIGKRLANLLANLENFGANADDFHLLGISLGAHIAGWAGKYFRLYKRRSIGRITGLDPAGPCFSHAYSDQRLDKSDALYVDVLHSNRLVQGIIEPLGHADFYINGGGPNQPGCMMPSCSHLRAAQVYTESVTTPKSFVGIRCQSWKHFEANACEKNMYAVLGYGSSTTSRGLYYLRTSGSPPFGLGMEGTKIVGPTFESWLRTLNIA
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Uniprot
H9JWY8
A0A2A4JGD9
A0A2H1X3Q9
A0A0L7KP71
A0A194PSE8
A0A194RQL7
+ More
A0A3S2L3L9 I6TFN4 A0A220K8L9 A0A2H1WNR8 A0A2H1W7F1 A0A3S2NZZ7 A0A2H1W795 A0A0L7L8R4 A0A2H1W7Q1 A0A2W1BV63 A0A2W1BXY3 E2C0E1 A0A2A4J5Z0 F4WY21 A0A0P4VMV4 A0A026X3P3 E9JDE9 A0A158NYL0 A0A194QC74 A0A146LFH2 A0A2A4IT83 E9JDF3 K7IZY1 E2A4Y7 A0A0A9XTP0 A0A1B6D221 A0A088AM94 A0A194QBZ0 A0A195FVA2 A0A1B6D5Q3 T1H9Y2 A0A195AZA7 A0A1B6I6C8 A0A0N1IGL3 K7IMY7 A0A232FGU7 A0A0A9XLC9 A0A067RM73 A0A0L0C2D6 A0A2A3EET9 E2BMN0 A0A1L8DQ87 A0A1L8DPY6 E9IHA8 K7IWE9 A0A087ZYV7 A0A195CR27 A0A151X5R7 A0A224XHV1 H9JQ12 Q16MG3 A0A3S2LBT4 A0A182TWL0 A0A2A4J4I9 A0A195BWG7 A0A158NEP4 A0A195FLQ2 A0A2H1WK50 A0A2H1WL24 A0A310STZ8 A0A1Y1NEG9 E0VJB4 A0A023ER10 A0A182GQ76 A0A0M3QTX4 A0A151XIN4 A0A023ER26 A0A182X488 A0A182URP2 A0A3F2YST1
A0A3S2L3L9 I6TFN4 A0A220K8L9 A0A2H1WNR8 A0A2H1W7F1 A0A3S2NZZ7 A0A2H1W795 A0A0L7L8R4 A0A2H1W7Q1 A0A2W1BV63 A0A2W1BXY3 E2C0E1 A0A2A4J5Z0 F4WY21 A0A0P4VMV4 A0A026X3P3 E9JDE9 A0A158NYL0 A0A194QC74 A0A146LFH2 A0A2A4IT83 E9JDF3 K7IZY1 E2A4Y7 A0A0A9XTP0 A0A1B6D221 A0A088AM94 A0A194QBZ0 A0A195FVA2 A0A1B6D5Q3 T1H9Y2 A0A195AZA7 A0A1B6I6C8 A0A0N1IGL3 K7IMY7 A0A232FGU7 A0A0A9XLC9 A0A067RM73 A0A0L0C2D6 A0A2A3EET9 E2BMN0 A0A1L8DQ87 A0A1L8DPY6 E9IHA8 K7IWE9 A0A087ZYV7 A0A195CR27 A0A151X5R7 A0A224XHV1 H9JQ12 Q16MG3 A0A3S2LBT4 A0A182TWL0 A0A2A4J4I9 A0A195BWG7 A0A158NEP4 A0A195FLQ2 A0A2H1WK50 A0A2H1WL24 A0A310STZ8 A0A1Y1NEG9 E0VJB4 A0A023ER10 A0A182GQ76 A0A0M3QTX4 A0A151XIN4 A0A023ER26 A0A182X488 A0A182URP2 A0A3F2YST1
Pubmed
EMBL
BABH01041781
NWSH01001670
PCG70462.1
ODYU01013233
SOQ59929.1
JTDY01007594
+ More
KOB65092.1 KQ459601 KPI94050.1 KQ459833 KPJ19782.1 RSAL01000184 RVE44850.1 JQ814807 AFM73623.1 MF319718 ASJ26432.1 ODYU01009953 SOQ54715.1 ODYU01006796 SOQ48977.1 RSAL01000012 RVE53468.1 SOQ48975.1 JTDY01002277 KOB71750.1 SOQ48976.1 KZ149926 PZC77545.1 PZC77546.1 GL451770 EFN78600.1 NWSH01003103 PCG66944.1 GL888434 EGI60946.1 GDKW01002938 JAI53657.1 KK107013 QOIP01000003 EZA62897.1 RLU25094.1 GL771866 EFZ09198.1 ADTU01004020 KQ459193 KPJ03024.1 GDHC01012863 JAQ05766.1 NWSH01007523 PCG62931.1 EFZ09320.1 GL436770 EFN71504.1 GBHO01020355 JAG23249.1 GEDC01024628 GEDC01017602 JAS12670.1 JAS19696.1 KPJ03068.1 KQ981276 KYN43794.1 GEDC01016286 JAS21012.1 ACPB03014206 KQ976701 KYM77372.1 GECU01025256 JAS82450.1 KQ460781 KPJ12226.1 NNAY01000216 OXU29964.1 GBHO01025674 GDHC01019063 JAG17930.1 JAP99565.1 KK852543 KDR21675.1 JRES01000987 KNC26392.1 KZ288269 PBC29994.1 GL449275 EFN83048.1 GFDF01005569 JAV08515.1 GFDF01005568 JAV08516.1 GL763196 EFZ20041.1 KQ977381 KYN03156.1 KQ982494 KYQ55731.1 GFTR01004361 JAW12065.1 BABH01012334 CH477862 EAT35525.1 RSAL01000052 RVE50204.1 NWSH01003437 PCG66302.1 KQ976401 KYM92313.1 ADTU01013601 KQ981490 KYN41261.1 ODYU01009136 SOQ53346.1 ODYU01009344 SOQ53707.1 KQ760495 OAD60121.1 GEZM01005476 JAV96149.1 DS235221 EEB13470.1 JXUM01153628 JXUM01153629 JXUM01153630 GAPW01002018 KQ571758 JAC11580.1 KXJ68117.1 JXUM01080013 JXUM01080014 KQ563159 KXJ74375.1 CP012523 ALC39639.1 KQ982080 KYQ60259.1 GAPW01002015 JAC11583.1 APCN01002931
KOB65092.1 KQ459601 KPI94050.1 KQ459833 KPJ19782.1 RSAL01000184 RVE44850.1 JQ814807 AFM73623.1 MF319718 ASJ26432.1 ODYU01009953 SOQ54715.1 ODYU01006796 SOQ48977.1 RSAL01000012 RVE53468.1 SOQ48975.1 JTDY01002277 KOB71750.1 SOQ48976.1 KZ149926 PZC77545.1 PZC77546.1 GL451770 EFN78600.1 NWSH01003103 PCG66944.1 GL888434 EGI60946.1 GDKW01002938 JAI53657.1 KK107013 QOIP01000003 EZA62897.1 RLU25094.1 GL771866 EFZ09198.1 ADTU01004020 KQ459193 KPJ03024.1 GDHC01012863 JAQ05766.1 NWSH01007523 PCG62931.1 EFZ09320.1 GL436770 EFN71504.1 GBHO01020355 JAG23249.1 GEDC01024628 GEDC01017602 JAS12670.1 JAS19696.1 KPJ03068.1 KQ981276 KYN43794.1 GEDC01016286 JAS21012.1 ACPB03014206 KQ976701 KYM77372.1 GECU01025256 JAS82450.1 KQ460781 KPJ12226.1 NNAY01000216 OXU29964.1 GBHO01025674 GDHC01019063 JAG17930.1 JAP99565.1 KK852543 KDR21675.1 JRES01000987 KNC26392.1 KZ288269 PBC29994.1 GL449275 EFN83048.1 GFDF01005569 JAV08515.1 GFDF01005568 JAV08516.1 GL763196 EFZ20041.1 KQ977381 KYN03156.1 KQ982494 KYQ55731.1 GFTR01004361 JAW12065.1 BABH01012334 CH477862 EAT35525.1 RSAL01000052 RVE50204.1 NWSH01003437 PCG66302.1 KQ976401 KYM92313.1 ADTU01013601 KQ981490 KYN41261.1 ODYU01009136 SOQ53346.1 ODYU01009344 SOQ53707.1 KQ760495 OAD60121.1 GEZM01005476 JAV96149.1 DS235221 EEB13470.1 JXUM01153628 JXUM01153629 JXUM01153630 GAPW01002018 KQ571758 JAC11580.1 KXJ68117.1 JXUM01080013 JXUM01080014 KQ563159 KXJ74375.1 CP012523 ALC39639.1 KQ982080 KYQ60259.1 GAPW01002015 JAC11583.1 APCN01002931
Proteomes
UP000005204
UP000218220
UP000037510
UP000053268
UP000053240
UP000283053
+ More
UP000008237 UP000007755 UP000053097 UP000279307 UP000005205 UP000002358 UP000000311 UP000005203 UP000078541 UP000015103 UP000078540 UP000215335 UP000027135 UP000037069 UP000242457 UP000078542 UP000075809 UP000008820 UP000075902 UP000009046 UP000069940 UP000249989 UP000092553 UP000076407 UP000075903 UP000075840
UP000008237 UP000007755 UP000053097 UP000279307 UP000005205 UP000002358 UP000000311 UP000005203 UP000078541 UP000015103 UP000078540 UP000215335 UP000027135 UP000037069 UP000242457 UP000078542 UP000075809 UP000008820 UP000075902 UP000009046 UP000069940 UP000249989 UP000092553 UP000076407 UP000075903 UP000075840
Pfam
PF00151 Lipase
Interpro
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
H9JWY8
A0A2A4JGD9
A0A2H1X3Q9
A0A0L7KP71
A0A194PSE8
A0A194RQL7
+ More
A0A3S2L3L9 I6TFN4 A0A220K8L9 A0A2H1WNR8 A0A2H1W7F1 A0A3S2NZZ7 A0A2H1W795 A0A0L7L8R4 A0A2H1W7Q1 A0A2W1BV63 A0A2W1BXY3 E2C0E1 A0A2A4J5Z0 F4WY21 A0A0P4VMV4 A0A026X3P3 E9JDE9 A0A158NYL0 A0A194QC74 A0A146LFH2 A0A2A4IT83 E9JDF3 K7IZY1 E2A4Y7 A0A0A9XTP0 A0A1B6D221 A0A088AM94 A0A194QBZ0 A0A195FVA2 A0A1B6D5Q3 T1H9Y2 A0A195AZA7 A0A1B6I6C8 A0A0N1IGL3 K7IMY7 A0A232FGU7 A0A0A9XLC9 A0A067RM73 A0A0L0C2D6 A0A2A3EET9 E2BMN0 A0A1L8DQ87 A0A1L8DPY6 E9IHA8 K7IWE9 A0A087ZYV7 A0A195CR27 A0A151X5R7 A0A224XHV1 H9JQ12 Q16MG3 A0A3S2LBT4 A0A182TWL0 A0A2A4J4I9 A0A195BWG7 A0A158NEP4 A0A195FLQ2 A0A2H1WK50 A0A2H1WL24 A0A310STZ8 A0A1Y1NEG9 E0VJB4 A0A023ER10 A0A182GQ76 A0A0M3QTX4 A0A151XIN4 A0A023ER26 A0A182X488 A0A182URP2 A0A3F2YST1
A0A3S2L3L9 I6TFN4 A0A220K8L9 A0A2H1WNR8 A0A2H1W7F1 A0A3S2NZZ7 A0A2H1W795 A0A0L7L8R4 A0A2H1W7Q1 A0A2W1BV63 A0A2W1BXY3 E2C0E1 A0A2A4J5Z0 F4WY21 A0A0P4VMV4 A0A026X3P3 E9JDE9 A0A158NYL0 A0A194QC74 A0A146LFH2 A0A2A4IT83 E9JDF3 K7IZY1 E2A4Y7 A0A0A9XTP0 A0A1B6D221 A0A088AM94 A0A194QBZ0 A0A195FVA2 A0A1B6D5Q3 T1H9Y2 A0A195AZA7 A0A1B6I6C8 A0A0N1IGL3 K7IMY7 A0A232FGU7 A0A0A9XLC9 A0A067RM73 A0A0L0C2D6 A0A2A3EET9 E2BMN0 A0A1L8DQ87 A0A1L8DPY6 E9IHA8 K7IWE9 A0A087ZYV7 A0A195CR27 A0A151X5R7 A0A224XHV1 H9JQ12 Q16MG3 A0A3S2LBT4 A0A182TWL0 A0A2A4J4I9 A0A195BWG7 A0A158NEP4 A0A195FLQ2 A0A2H1WK50 A0A2H1WL24 A0A310STZ8 A0A1Y1NEG9 E0VJB4 A0A023ER10 A0A182GQ76 A0A0M3QTX4 A0A151XIN4 A0A023ER26 A0A182X488 A0A182URP2 A0A3F2YST1
PDB
1W52
E-value=2.18801e-33,
Score=355
Ontologies
GO
PANTHER
Topology
Subcellular location
Secreted
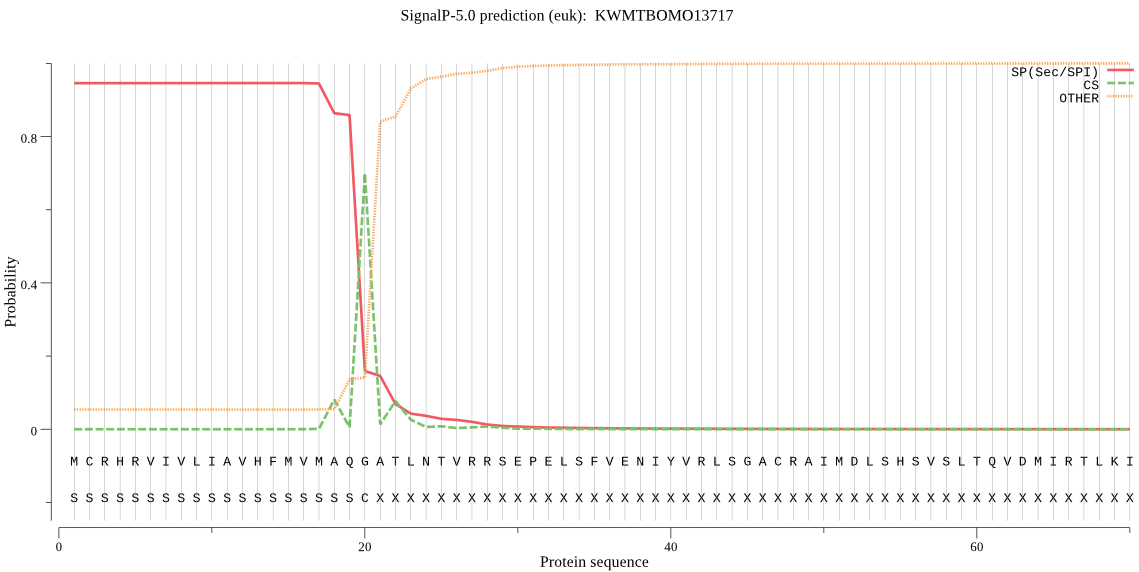
SignalP
Position: 1 - 20,
Likelihood: 0.945468
Length:
361
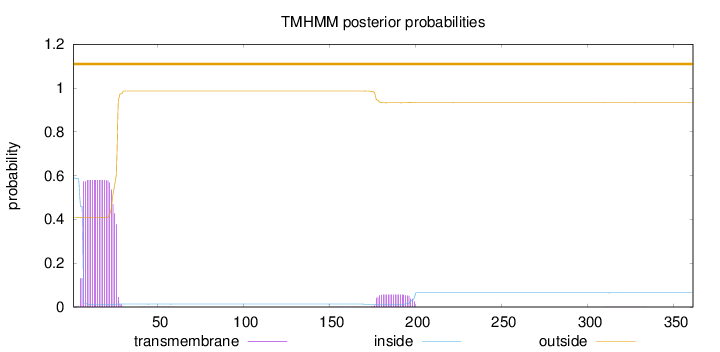
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
12.62855
Exp number, first 60 AAs:
11.40638
Total prob of N-in:
0.59025
POSSIBLE N-term signal
sequence
outside
1 - 361
Population Genetic Test Statistics
Pi
213.965328
Theta
219.460949
Tajima's D
-0.543032
CLR
383.951962
CSRT
0.231188440577971
Interpretation
Uncertain
