Gene
KWMTBOMO13716
Annotation
PREDICTED:_D-beta-hydroxybutyrate_dehydrogenase?_mitochondrial-like_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 2.281
Sequence
CDS
ATGGTTGCCGGGGACGTAACTCGGCGAGCGTCGATCACGGCGCCGTCGATGCATCGTCGGCCTTCAGCACGTCGAGGCTCCCTGATTAAAGGCCCATCGAACCCGTCTCAGGAAGTTCCATGGGATATCATCGACCGCTGTGCACTGCCCATTGTCTTATGCCACGCCCTCGCCGTCGTACTCTCCACGATCCTGAACTCCCTGCATATCAGTCAAGTGTCGGTCTTCACGCTATTTTTATGGTTTGCAATTTCCGTAACTGGATCCTTGTGGTTCTACCATAATTTACAGGTAACGGCTGCCGGCAAAGCAGTACTTGTCACTGGGTGTGACAATGTATTAGGCAACGCACTGGCCCGAAGACTGGACGATATGGGATATCATGTGTTTGCTGGCTTTCAAACTAAAGCGGGCAACATCGATGCAGATATGCTGAAAGAAGATTGCTCTGGAAGATTACATACCCTACAACTTGACATTACGTCTGAAACCCAGATCCTATCGGCCTCCTTGTACATCGTGGACCATCTACCAGAGGGCGCCCAGGGTTTGTGGGCAATTATTAACTGCGAGTGTTGGTGCGCTTTAGGTGAACTTGAATGGGTTCCCTTTTCCGTTATCAAACGATCCATGGATGTCAACCTTTTGGGTCCAGCTCGTCTGGTGCAAGTGATGCTGCCTCTGGTACGACGTGCTCGCGGTCGTATTGTACTGGCTTCCTCCATTTTAACGCACGTCGCGGCCCCTGTACGCGGGGTCCACGCCGCTTCTCTCGCAGCCCTCGACGCTATGGCTGCCTGTTTGCGTCGTGAGCTCAAGCCACGAGGTGTCGATGTTGTGGTAGTGGCCGCTGGAGAATATACGACAGGTAGCGCTTGGTTATCCGAAGAGAAACTGCTGGAACAAGCCCGTGAGATGTGGAAACGACTCAGTGATGAACAGAAAGGAGCTTATGGCGAAGACTACTTCGAGCAGGCTCTCCGAAGCCTCGAAAAATACACTAAAAGCGCTGATGCCGATTTATCAGCGGTTACTCGCGCTCTAAGTGACGGTGTGACACGTACGTTCCCGCTGTCTCGCTACACGCCGGTCTCTCCAAGGGAGAAACTGAAGTCTCTACTCGCAGAGCACATGCCACGCTCCCTCTACGAAGGTCTATACACAGATTAA
Protein
MVAGDVTRRASITAPSMHRRPSARRGSLIKGPSNPSQEVPWDIIDRCALPIVLCHALAVVLSTILNSLHISQVSVFTLFLWFAISVTGSLWFYHNLQVTAAGKAVLVTGCDNVLGNALARRLDDMGYHVFAGFQTKAGNIDADMLKEDCSGRLHTLQLDITSETQILSASLYIVDHLPEGAQGLWAIINCECWCALGELEWVPFSVIKRSMDVNLLGPARLVQVMLPLVRRARGRIVLASSILTHVAAPVRGVHAASLAALDAMAACLRRELKPRGVDVVVVAAGEYTTGSAWLSEEKLLEQAREMWKRLSDEQKGAYGEDYFEQALRSLEKYTKSADADLSAVTRALSDGVTRTFPLSRYTPVSPREKLKSLLAEHMPRSLYEGLYTD
Summary
Uniprot
A0A2A4JET5
A0A194RUG9
A0A3S2LVF5
A0A194PKT9
A0A212FJN7
A0A2H1VRF7
+ More
D6WA35 A0A1Y1KAB9 A0A1W4WEA4 A0A1L8E4L0 A0A336M7P3 A0A023ESE1 A0A067RU64 A0A1Q3FZK8 A0A2J7RD87 A0A1Q3FZL9 A0A1Q3FZQ2 D3TMP5 A0A0L7LA14 A0A1I8P1D2 T1PA61 A0A1B0G738 A0A1I8N6K1 A0A034WW19 A0A0P9ACJ5 B3MH34 W8C8H7 B4J5G3 B4MR19 A0A0K8W4G6 B4KPV2 A0A0A1XTC3 Q28ZV9 A0A3B0JW03 A0A1W4ULW6 A0A0J9RBM1 A0A0M3QVD1 B3NS88 Q7K3N4 B4P5P8 A0A026VVA3 E2C6N5 A0A0L7QL15 A0A1B6K4I1 A0A1B6GCV4 A0A1Q3FZJ9 A0A1Q3FZN9 A0A1Q3FZR5 F4WHK6 A0A151K124 A0A151J580 A0A158N901 A0A1B6CSP8 A0A151IBL0 A0A151I124 E2A1G5 A0A0J7L4L6 A0A0C9Q5B7 A0A0L0C680 A0A154PLC8 A0A2A3ER37 A0A088AV05 A0A1B6LI94 Q16M72 A0A151WNJ4 A0A1B0CMC6 A0A1J1HMD6 E0VWY3 A0A0K8WEJ4 A0A182IXC3 A0A084WJ72 A0A2J7RD83 A0A182FUN5 A0A1A9WQX0 A0A1B0AG70 A0A1A9UVK8 A0A0N1IT64 A0A182QA96 A0A182XGQ1 A0A182HHB7 A0NGR6 A0A182VA18 A0A182YL79 A0A182L4U1 A0A182P527 A0A1A9XIC4 A0A1B0BUD6 A0A182NSZ9 A0A182R7G3 B4HNX4 B4LKV0 A0A182MGU7 A0A182ULE5 A0A023F8F1 A0A0A9XQ23 T1HIH0 A0A2P8Y1C1 R4FN19
D6WA35 A0A1Y1KAB9 A0A1W4WEA4 A0A1L8E4L0 A0A336M7P3 A0A023ESE1 A0A067RU64 A0A1Q3FZK8 A0A2J7RD87 A0A1Q3FZL9 A0A1Q3FZQ2 D3TMP5 A0A0L7LA14 A0A1I8P1D2 T1PA61 A0A1B0G738 A0A1I8N6K1 A0A034WW19 A0A0P9ACJ5 B3MH34 W8C8H7 B4J5G3 B4MR19 A0A0K8W4G6 B4KPV2 A0A0A1XTC3 Q28ZV9 A0A3B0JW03 A0A1W4ULW6 A0A0J9RBM1 A0A0M3QVD1 B3NS88 Q7K3N4 B4P5P8 A0A026VVA3 E2C6N5 A0A0L7QL15 A0A1B6K4I1 A0A1B6GCV4 A0A1Q3FZJ9 A0A1Q3FZN9 A0A1Q3FZR5 F4WHK6 A0A151K124 A0A151J580 A0A158N901 A0A1B6CSP8 A0A151IBL0 A0A151I124 E2A1G5 A0A0J7L4L6 A0A0C9Q5B7 A0A0L0C680 A0A154PLC8 A0A2A3ER37 A0A088AV05 A0A1B6LI94 Q16M72 A0A151WNJ4 A0A1B0CMC6 A0A1J1HMD6 E0VWY3 A0A0K8WEJ4 A0A182IXC3 A0A084WJ72 A0A2J7RD83 A0A182FUN5 A0A1A9WQX0 A0A1B0AG70 A0A1A9UVK8 A0A0N1IT64 A0A182QA96 A0A182XGQ1 A0A182HHB7 A0NGR6 A0A182VA18 A0A182YL79 A0A182L4U1 A0A182P527 A0A1A9XIC4 A0A1B0BUD6 A0A182NSZ9 A0A182R7G3 B4HNX4 B4LKV0 A0A182MGU7 A0A182ULE5 A0A023F8F1 A0A0A9XQ23 T1HIH0 A0A2P8Y1C1 R4FN19
Pubmed
26354079
22118469
18362917
19820115
28004739
24945155
+ More
24845553 20353571 26227816 25315136 25348373 17994087 18057021 24495485 25830018 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24508170 30249741 20798317 21719571 21347285 26108605 17510324 20566863 24438588 12364791 25244985 20966253 25474469 25401762 26823975 29403074
24845553 20353571 26227816 25315136 25348373 17994087 18057021 24495485 25830018 15632085 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 24508170 30249741 20798317 21719571 21347285 26108605 17510324 20566863 24438588 12364791 25244985 20966253 25474469 25401762 26823975 29403074
EMBL
NWSH01001670
PCG70461.1
KQ459833
KPJ19781.1
RSAL01000184
RVE44848.1
+ More
KQ459601 KPI94051.1 AGBW02008253 OWR53920.1 ODYU01003973 SOQ43378.1 KQ971312 EEZ98067.1 GEZM01088099 JAV58314.1 GFDF01000520 JAV13564.1 UFQT01000226 SSX22058.1 GAPW01001927 JAC11671.1 KK852463 KDR23384.1 GFDL01002068 JAV32977.1 NEVH01005293 PNF38795.1 GFDL01002075 JAV32970.1 GFDL01002032 JAV33013.1 EZ422697 ADD18973.1 JTDY01002053 KOB72229.1 KA644828 AFP59457.1 CCAG010003773 CCAG010003774 GAKP01000954 JAC57998.1 CH902619 KPU75824.1 EDV35793.1 KPU75825.1 GAMC01007457 GAMC01007456 JAB99099.1 CH916367 EDW00726.1 CH963849 EDW74558.1 GDHF01006574 JAI45740.1 CH933808 EDW10229.1 KRG05097.1 GBXI01000424 JAD13868.1 CM000071 EAL25503.2 OUUW01000001 SPP75248.1 CM002911 KMY93089.1 CP012524 ALC42209.1 CH954179 EDV56390.1 AE013599 AY058459 AAF58586.1 AAL13688.1 CM000158 EDW90845.1 KK108326 QOIP01000004 EZA46759.1 RLU23365.1 GL453160 EFN76437.1 KQ414934 KOC59285.1 GECU01001613 JAT06094.1 GECZ01009513 JAS60256.1 GFDL01002060 JAV32985.1 GFDL01002055 JAV32990.1 GFDL01002008 JAV33037.1 GL888161 EGI66360.1 KQ981229 KYN44345.1 KQ980063 KYN17958.1 ADTU01009011 GEDC01025630 GEDC01020901 JAS11668.1 JAS16397.1 KQ978095 KYM97027.1 KQ976581 KYM79879.1 GL435766 EFN72638.1 LBMM01000785 KMQ97498.1 GBYB01009243 JAG79010.1 JRES01000848 KNC27762.1 KQ434954 KZC12557.1 KZ288192 PBC34198.1 GEBQ01016607 JAT23370.1 CH477874 EAT35431.1 KQ982907 KYQ49449.1 AJWK01018572 AJWK01018573 AJWK01018574 CVRI01000011 CRK89215.1 DS235824 EEB17889.1 GDHF01002792 JAI49522.1 ATLV01023975 KE525347 KFB50266.1 PNF38794.1 KQ435883 KOX69724.1 AXCN02002127 APCN01002400 APCN01002401 AAAB01008986 EAU75862.2 JXJN01020619 CH480816 EDW47489.1 CH940648 EDW60754.2 AXCM01005637 GBBI01001086 JAC17626.1 GBHO01037661 GBHO01033629 GBHO01025877 GBHO01020766 GBHO01020763 GBHO01020762 GBRD01008736 GDHC01002310 JAG05943.1 JAG09975.1 JAG17727.1 JAG22838.1 JAG22841.1 JAG22842.1 JAG57085.1 JAQ16319.1 ACPB03001249 PYGN01001052 PSN38057.1 GAHY01001499 JAA76011.1
KQ459601 KPI94051.1 AGBW02008253 OWR53920.1 ODYU01003973 SOQ43378.1 KQ971312 EEZ98067.1 GEZM01088099 JAV58314.1 GFDF01000520 JAV13564.1 UFQT01000226 SSX22058.1 GAPW01001927 JAC11671.1 KK852463 KDR23384.1 GFDL01002068 JAV32977.1 NEVH01005293 PNF38795.1 GFDL01002075 JAV32970.1 GFDL01002032 JAV33013.1 EZ422697 ADD18973.1 JTDY01002053 KOB72229.1 KA644828 AFP59457.1 CCAG010003773 CCAG010003774 GAKP01000954 JAC57998.1 CH902619 KPU75824.1 EDV35793.1 KPU75825.1 GAMC01007457 GAMC01007456 JAB99099.1 CH916367 EDW00726.1 CH963849 EDW74558.1 GDHF01006574 JAI45740.1 CH933808 EDW10229.1 KRG05097.1 GBXI01000424 JAD13868.1 CM000071 EAL25503.2 OUUW01000001 SPP75248.1 CM002911 KMY93089.1 CP012524 ALC42209.1 CH954179 EDV56390.1 AE013599 AY058459 AAF58586.1 AAL13688.1 CM000158 EDW90845.1 KK108326 QOIP01000004 EZA46759.1 RLU23365.1 GL453160 EFN76437.1 KQ414934 KOC59285.1 GECU01001613 JAT06094.1 GECZ01009513 JAS60256.1 GFDL01002060 JAV32985.1 GFDL01002055 JAV32990.1 GFDL01002008 JAV33037.1 GL888161 EGI66360.1 KQ981229 KYN44345.1 KQ980063 KYN17958.1 ADTU01009011 GEDC01025630 GEDC01020901 JAS11668.1 JAS16397.1 KQ978095 KYM97027.1 KQ976581 KYM79879.1 GL435766 EFN72638.1 LBMM01000785 KMQ97498.1 GBYB01009243 JAG79010.1 JRES01000848 KNC27762.1 KQ434954 KZC12557.1 KZ288192 PBC34198.1 GEBQ01016607 JAT23370.1 CH477874 EAT35431.1 KQ982907 KYQ49449.1 AJWK01018572 AJWK01018573 AJWK01018574 CVRI01000011 CRK89215.1 DS235824 EEB17889.1 GDHF01002792 JAI49522.1 ATLV01023975 KE525347 KFB50266.1 PNF38794.1 KQ435883 KOX69724.1 AXCN02002127 APCN01002400 APCN01002401 AAAB01008986 EAU75862.2 JXJN01020619 CH480816 EDW47489.1 CH940648 EDW60754.2 AXCM01005637 GBBI01001086 JAC17626.1 GBHO01037661 GBHO01033629 GBHO01025877 GBHO01020766 GBHO01020763 GBHO01020762 GBRD01008736 GDHC01002310 JAG05943.1 JAG09975.1 JAG17727.1 JAG22838.1 JAG22841.1 JAG22842.1 JAG57085.1 JAQ16319.1 ACPB03001249 PYGN01001052 PSN38057.1 GAHY01001499 JAA76011.1
Proteomes
UP000218220
UP000053240
UP000283053
UP000053268
UP000007151
UP000007266
+ More
UP000192223 UP000027135 UP000235965 UP000037510 UP000095300 UP000092444 UP000095301 UP000007801 UP000001070 UP000007798 UP000009192 UP000001819 UP000268350 UP000192221 UP000092553 UP000008711 UP000000803 UP000002282 UP000053097 UP000279307 UP000008237 UP000053825 UP000007755 UP000078541 UP000078492 UP000005205 UP000078542 UP000078540 UP000000311 UP000036403 UP000037069 UP000076502 UP000242457 UP000005203 UP000008820 UP000075809 UP000092461 UP000183832 UP000009046 UP000075880 UP000030765 UP000069272 UP000091820 UP000092445 UP000078200 UP000053105 UP000075886 UP000076407 UP000075840 UP000007062 UP000075903 UP000076408 UP000075882 UP000075885 UP000092443 UP000092460 UP000075884 UP000075900 UP000001292 UP000008792 UP000075883 UP000075902 UP000015103 UP000245037
UP000192223 UP000027135 UP000235965 UP000037510 UP000095300 UP000092444 UP000095301 UP000007801 UP000001070 UP000007798 UP000009192 UP000001819 UP000268350 UP000192221 UP000092553 UP000008711 UP000000803 UP000002282 UP000053097 UP000279307 UP000008237 UP000053825 UP000007755 UP000078541 UP000078492 UP000005205 UP000078542 UP000078540 UP000000311 UP000036403 UP000037069 UP000076502 UP000242457 UP000005203 UP000008820 UP000075809 UP000092461 UP000183832 UP000009046 UP000075880 UP000030765 UP000069272 UP000091820 UP000092445 UP000078200 UP000053105 UP000075886 UP000076407 UP000075840 UP000007062 UP000075903 UP000076408 UP000075882 UP000075885 UP000092443 UP000092460 UP000075884 UP000075900 UP000001292 UP000008792 UP000075883 UP000075902 UP000015103 UP000245037
PRIDE
Pfam
PF00106 adh_short
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
A0A2A4JET5
A0A194RUG9
A0A3S2LVF5
A0A194PKT9
A0A212FJN7
A0A2H1VRF7
+ More
D6WA35 A0A1Y1KAB9 A0A1W4WEA4 A0A1L8E4L0 A0A336M7P3 A0A023ESE1 A0A067RU64 A0A1Q3FZK8 A0A2J7RD87 A0A1Q3FZL9 A0A1Q3FZQ2 D3TMP5 A0A0L7LA14 A0A1I8P1D2 T1PA61 A0A1B0G738 A0A1I8N6K1 A0A034WW19 A0A0P9ACJ5 B3MH34 W8C8H7 B4J5G3 B4MR19 A0A0K8W4G6 B4KPV2 A0A0A1XTC3 Q28ZV9 A0A3B0JW03 A0A1W4ULW6 A0A0J9RBM1 A0A0M3QVD1 B3NS88 Q7K3N4 B4P5P8 A0A026VVA3 E2C6N5 A0A0L7QL15 A0A1B6K4I1 A0A1B6GCV4 A0A1Q3FZJ9 A0A1Q3FZN9 A0A1Q3FZR5 F4WHK6 A0A151K124 A0A151J580 A0A158N901 A0A1B6CSP8 A0A151IBL0 A0A151I124 E2A1G5 A0A0J7L4L6 A0A0C9Q5B7 A0A0L0C680 A0A154PLC8 A0A2A3ER37 A0A088AV05 A0A1B6LI94 Q16M72 A0A151WNJ4 A0A1B0CMC6 A0A1J1HMD6 E0VWY3 A0A0K8WEJ4 A0A182IXC3 A0A084WJ72 A0A2J7RD83 A0A182FUN5 A0A1A9WQX0 A0A1B0AG70 A0A1A9UVK8 A0A0N1IT64 A0A182QA96 A0A182XGQ1 A0A182HHB7 A0NGR6 A0A182VA18 A0A182YL79 A0A182L4U1 A0A182P527 A0A1A9XIC4 A0A1B0BUD6 A0A182NSZ9 A0A182R7G3 B4HNX4 B4LKV0 A0A182MGU7 A0A182ULE5 A0A023F8F1 A0A0A9XQ23 T1HIH0 A0A2P8Y1C1 R4FN19
D6WA35 A0A1Y1KAB9 A0A1W4WEA4 A0A1L8E4L0 A0A336M7P3 A0A023ESE1 A0A067RU64 A0A1Q3FZK8 A0A2J7RD87 A0A1Q3FZL9 A0A1Q3FZQ2 D3TMP5 A0A0L7LA14 A0A1I8P1D2 T1PA61 A0A1B0G738 A0A1I8N6K1 A0A034WW19 A0A0P9ACJ5 B3MH34 W8C8H7 B4J5G3 B4MR19 A0A0K8W4G6 B4KPV2 A0A0A1XTC3 Q28ZV9 A0A3B0JW03 A0A1W4ULW6 A0A0J9RBM1 A0A0M3QVD1 B3NS88 Q7K3N4 B4P5P8 A0A026VVA3 E2C6N5 A0A0L7QL15 A0A1B6K4I1 A0A1B6GCV4 A0A1Q3FZJ9 A0A1Q3FZN9 A0A1Q3FZR5 F4WHK6 A0A151K124 A0A151J580 A0A158N901 A0A1B6CSP8 A0A151IBL0 A0A151I124 E2A1G5 A0A0J7L4L6 A0A0C9Q5B7 A0A0L0C680 A0A154PLC8 A0A2A3ER37 A0A088AV05 A0A1B6LI94 Q16M72 A0A151WNJ4 A0A1B0CMC6 A0A1J1HMD6 E0VWY3 A0A0K8WEJ4 A0A182IXC3 A0A084WJ72 A0A2J7RD83 A0A182FUN5 A0A1A9WQX0 A0A1B0AG70 A0A1A9UVK8 A0A0N1IT64 A0A182QA96 A0A182XGQ1 A0A182HHB7 A0NGR6 A0A182VA18 A0A182YL79 A0A182L4U1 A0A182P527 A0A1A9XIC4 A0A1B0BUD6 A0A182NSZ9 A0A182R7G3 B4HNX4 B4LKV0 A0A182MGU7 A0A182ULE5 A0A023F8F1 A0A0A9XQ23 T1HIH0 A0A2P8Y1C1 R4FN19
PDB
4OSO
E-value=9.08166e-07,
Score=126
Ontologies
Topology
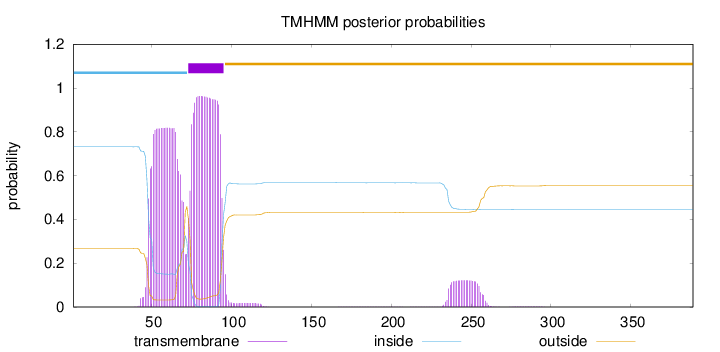
Length:
389
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
40.49584
Exp number, first 60 AAs:
10.35118
Total prob of N-in:
0.73165
POSSIBLE N-term signal
sequence
inside
1 - 72
TMhelix
73 - 95
outside
96 - 389
Population Genetic Test Statistics
Pi
265.213189
Theta
221.838129
Tajima's D
-2.641935
CLR
10.8581
CSRT
0.000199990000499975
Interpretation
Possibly Positive selection
