Gene
KWMTBOMO13715
Pre Gene Modal
BGIBMGA011008
Annotation
cell_death_activator_CIDE-B_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.692
Sequence
CDS
ATGGAAAACGACATTAACAAACCATATAAAATTTGCGATGTTAATCGCGAAAAGAAGAAAGGAATTGTCGCGATGTCGCTTGAAGACTTACTCATAAAAGTGCCGGAAAAGCTGGGAATGCCTTCTGAAAATTTAACTGTTGTATTGGAATCAGATGGTACAGAAGTTGATGATGAAGAATACTTCTCTACGTTAGACCCAGACACGTCCCTCATGATTTTGCACGGAAATGAAAAGTGGGTCCCGAACATGCCTAAGTGTCAGGTATCGTTAGATCAGACCGATGATATAGCTTTGGGTGATAAAGAACAAGTGGCGAGTCTTGTTGGTCGACTGCAGCATAACCTATGTCACATCTCGTTGCTTGGAGGCCAAGACCTAGAATTGTTATCTGATATGGATCCTGACAGCTTGGCTGACATTGTTACTGATCGAGATAATAGAATAATTTTGGAACATATTAAAGAAGCATCTGGAAGAATTCTTCTGGAAAAACGTCAAGCCCAAGATGCTATGGAATTGCTAAAATTATACCATCAGAGTGTCTCCAATGGTCCAGAAGACGTGTCTTCCCCAAAGCAGGAGAGGGTTTAA
Protein
MENDINKPYKICDVNREKKKGIVAMSLEDLLIKVPEKLGMPSENLTVVLESDGTEVDDEEYFSTLDPDTSLMILHGNEKWVPNMPKCQVSLDQTDDIALGDKEQVASLVGRLQHNLCHISLLGGQDLELLSDMDPDSLADIVTDRDNRIILEHIKEASGRILLEKRQAQDAMELLKLYHQSVSNGPEDVSSPKQERV
Summary
Uniprot
B0FC96
A0A2H1W0V8
D3JSV7
A0A1E1W5N0
A0A2A4JGA6
A0A194PN41
+ More
S4P8N0 A0A3S2N966 A0A194QMM7 A0A212FJN3 H9JNA5 A0A0L7KP39 A0A1Y1M327 A0A1W4WQE4 J3JZH4 D6WA34 A0A2J7RD86 N6TRH6 A0A067RRT3 A0A2P8Y1C4 A0A2R7W5T4 A0A151IBY9 A0A0J7L3N9 A0A2S2N9B7 A0A0L7QL78 E2A1G4 F4WHK7 A0A151I114 A0A151K1N2 A0A158N900 A0A151J514 A0A2H8TSM0 A0A026VSY1 A0A151WNQ8 E9JAU5 J9JNI3 E2C6N4 A0A2A3ER65 V9IHW8 A0A088AUR4 A0A1J1HMP5 A0A1B6MIX1 A0A1B6GC46 R4G377 A0A1B6DP76 A0A0A9XJA2 A0A1S3CVE3 A0A1Q3EVC5 A0A154PMN3 E0VWD7 A0A182QWK9 A0A182HFQ6 A0A1J1HME3 A0A1I8N5U7 A0A1S4G3C0 B0WG49 A0A0L0C8P1 A0A1J1HRV6 W5J8V0 A0A182P8G5 A0A1I8PYT9 A0A1B0G737 Q16FC1 A0A182MYE3 A0A182JPX1 A0A182J411 A0A182RLP3 A0A182SCB7 A0A182U9F5 A0A182X1U1 A0A182LQ35 A0A0M8ZRN7 A0A182WCU8 A0A182VLQ8 B4KPU8 B4LKV3 A0A182YA41 A0A0A1WNW0 A0A084WI53 A0A034WGY6 A0A023VTR5 A0A0K8U151 A0A1A9XIC9 W8ATX3 A0A224XZA8 B4J5G5 A0A1B0AG69 B4QC70 A0A336MMA2 K1RJK1 A0A1A9WQX9 A0A1S3DKZ7 B3NS92 Q6NR36 B4HNX1 A0A1W4U9Y1 B4P5Q2 A0A1B0BUD3 A0A0B4KFQ3
S4P8N0 A0A3S2N966 A0A194QMM7 A0A212FJN3 H9JNA5 A0A0L7KP39 A0A1Y1M327 A0A1W4WQE4 J3JZH4 D6WA34 A0A2J7RD86 N6TRH6 A0A067RRT3 A0A2P8Y1C4 A0A2R7W5T4 A0A151IBY9 A0A0J7L3N9 A0A2S2N9B7 A0A0L7QL78 E2A1G4 F4WHK7 A0A151I114 A0A151K1N2 A0A158N900 A0A151J514 A0A2H8TSM0 A0A026VSY1 A0A151WNQ8 E9JAU5 J9JNI3 E2C6N4 A0A2A3ER65 V9IHW8 A0A088AUR4 A0A1J1HMP5 A0A1B6MIX1 A0A1B6GC46 R4G377 A0A1B6DP76 A0A0A9XJA2 A0A1S3CVE3 A0A1Q3EVC5 A0A154PMN3 E0VWD7 A0A182QWK9 A0A182HFQ6 A0A1J1HME3 A0A1I8N5U7 A0A1S4G3C0 B0WG49 A0A0L0C8P1 A0A1J1HRV6 W5J8V0 A0A182P8G5 A0A1I8PYT9 A0A1B0G737 Q16FC1 A0A182MYE3 A0A182JPX1 A0A182J411 A0A182RLP3 A0A182SCB7 A0A182U9F5 A0A182X1U1 A0A182LQ35 A0A0M8ZRN7 A0A182WCU8 A0A182VLQ8 B4KPU8 B4LKV3 A0A182YA41 A0A0A1WNW0 A0A084WI53 A0A034WGY6 A0A023VTR5 A0A0K8U151 A0A1A9XIC9 W8ATX3 A0A224XZA8 B4J5G5 A0A1B0AG69 B4QC70 A0A336MMA2 K1RJK1 A0A1A9WQX9 A0A1S3DKZ7 B3NS92 Q6NR36 B4HNX1 A0A1W4U9Y1 B4P5Q2 A0A1B0BUD3 A0A0B4KFQ3
Pubmed
26354079
23622113
22118469
19121390
26227816
28004739
+ More
22516182 23537049 18362917 19820115 24845553 29403074 20798317 21719571 21347285 24508170 30249741 21282665 25401762 26823975 20566863 25315136 17510324 26108605 20920257 23761445 20966253 17994087 25244985 25830018 24438588 25348373 24642253 24495485 22936249 22992520 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
22516182 23537049 18362917 19820115 24845553 29403074 20798317 21719571 21347285 24508170 30249741 21282665 25401762 26823975 20566863 25315136 17510324 26108605 20920257 23761445 20966253 17994087 25244985 25830018 24438588 25348373 24642253 24495485 22936249 22992520 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304
EMBL
EU305741
ABY50540.1
ODYU01005373
SOQ46144.1
GU288814
ADB78701.1
+ More
GDQN01008767 JAT82287.1 NWSH01001670 PCG70460.1 KQ459600 KPI94154.1 GAIX01009405 JAA83155.1 RSAL01000184 RVE44847.1 KQ461195 KPJ06777.1 AGBW02008253 OWR53919.1 BABH01019629 JTDY01008133 KOB64719.1 GEZM01042406 JAV79881.1 BT128656 KB632391 AEE63613.1 ERL94534.1 KQ971312 EEZ98068.1 NEVH01005293 PNF38799.1 APGK01022561 KB740432 ENN80663.1 KK852463 KDR23385.1 PYGN01001052 PSN38058.1 KK854364 PTY15096.1 KQ978095 KYM97028.1 LBMM01000785 KMQ97497.1 GGMR01000913 MBY13532.1 KQ414934 KOC59286.1 GL435766 EFN72637.1 GL888161 EGI66361.1 KQ976581 KYM79878.1 KQ981229 KYN44346.1 ADTU01009011 KQ980063 KYN17959.1 GFXV01005382 MBW17187.1 KK108326 QOIP01000004 EZA46760.1 RLU23366.1 KQ982907 KYQ49448.1 GL770679 EFZ10044.1 ABLF02039438 GL453160 EFN76436.1 KZ288192 PBC34197.1 JR048336 AEY60635.1 CVRI01000011 CRK89211.1 GEBQ01004067 JAT35910.1 GECZ01009765 JAS60004.1 ACPB03001264 GAHY01001926 JAA75584.1 GEDC01027842 GEDC01020905 GEDC01009813 JAS09456.1 JAS16393.1 JAS27485.1 GBHO01022798 GBHO01022797 GDHC01012985 JAG20806.1 JAG20807.1 JAQ05644.1 GFDL01015781 JAV19264.1 KQ434954 KZC12558.1 DS235819 EEB17688.1 AXCN02001048 APCN01005563 APCN01005564 APCN01005565 CRK89213.1 DS231923 EDS26745.1 JRES01000848 KNC27764.1 CRK89214.1 ADMH02002040 ETN59808.1 CCAG010003771 CCAG010003772 CH478464 EAT32934.1 KQ435883 KOX69725.1 CH933808 EDW10225.1 CH940648 EDW60757.2 GBXI01013563 JAD00729.1 ATLV01023913 KE525347 KFB49897.1 GAKP01005370 JAC53582.1 KF745191 AHY19024.1 GDHF01032249 GDHF01006099 JAI20065.1 JAI46215.1 GAMC01016898 JAB89657.1 GFTR01002494 JAW13932.1 CH916367 EDW00728.1 CM000362 CM002911 EDX06701.1 KMY93082.1 UFQT01001192 SSX29417.1 JH816168 EKC41810.1 CH954179 EDV56394.2 AE013599 BT010245 AAF58589.1 AAQ23563.1 CH480816 EDW47486.1 CM000158 EDW90849.2 JXJN01020618 AGB93442.1
GDQN01008767 JAT82287.1 NWSH01001670 PCG70460.1 KQ459600 KPI94154.1 GAIX01009405 JAA83155.1 RSAL01000184 RVE44847.1 KQ461195 KPJ06777.1 AGBW02008253 OWR53919.1 BABH01019629 JTDY01008133 KOB64719.1 GEZM01042406 JAV79881.1 BT128656 KB632391 AEE63613.1 ERL94534.1 KQ971312 EEZ98068.1 NEVH01005293 PNF38799.1 APGK01022561 KB740432 ENN80663.1 KK852463 KDR23385.1 PYGN01001052 PSN38058.1 KK854364 PTY15096.1 KQ978095 KYM97028.1 LBMM01000785 KMQ97497.1 GGMR01000913 MBY13532.1 KQ414934 KOC59286.1 GL435766 EFN72637.1 GL888161 EGI66361.1 KQ976581 KYM79878.1 KQ981229 KYN44346.1 ADTU01009011 KQ980063 KYN17959.1 GFXV01005382 MBW17187.1 KK108326 QOIP01000004 EZA46760.1 RLU23366.1 KQ982907 KYQ49448.1 GL770679 EFZ10044.1 ABLF02039438 GL453160 EFN76436.1 KZ288192 PBC34197.1 JR048336 AEY60635.1 CVRI01000011 CRK89211.1 GEBQ01004067 JAT35910.1 GECZ01009765 JAS60004.1 ACPB03001264 GAHY01001926 JAA75584.1 GEDC01027842 GEDC01020905 GEDC01009813 JAS09456.1 JAS16393.1 JAS27485.1 GBHO01022798 GBHO01022797 GDHC01012985 JAG20806.1 JAG20807.1 JAQ05644.1 GFDL01015781 JAV19264.1 KQ434954 KZC12558.1 DS235819 EEB17688.1 AXCN02001048 APCN01005563 APCN01005564 APCN01005565 CRK89213.1 DS231923 EDS26745.1 JRES01000848 KNC27764.1 CRK89214.1 ADMH02002040 ETN59808.1 CCAG010003771 CCAG010003772 CH478464 EAT32934.1 KQ435883 KOX69725.1 CH933808 EDW10225.1 CH940648 EDW60757.2 GBXI01013563 JAD00729.1 ATLV01023913 KE525347 KFB49897.1 GAKP01005370 JAC53582.1 KF745191 AHY19024.1 GDHF01032249 GDHF01006099 JAI20065.1 JAI46215.1 GAMC01016898 JAB89657.1 GFTR01002494 JAW13932.1 CH916367 EDW00728.1 CM000362 CM002911 EDX06701.1 KMY93082.1 UFQT01001192 SSX29417.1 JH816168 EKC41810.1 CH954179 EDV56394.2 AE013599 BT010245 AAF58589.1 AAQ23563.1 CH480816 EDW47486.1 CM000158 EDW90849.2 JXJN01020618 AGB93442.1
Proteomes
UP000218220
UP000053268
UP000283053
UP000053240
UP000007151
UP000005204
+ More
UP000037510 UP000192223 UP000030742 UP000007266 UP000235965 UP000019118 UP000027135 UP000245037 UP000078542 UP000036403 UP000053825 UP000000311 UP000007755 UP000078540 UP000078541 UP000005205 UP000078492 UP000053097 UP000279307 UP000075809 UP000007819 UP000008237 UP000242457 UP000005203 UP000183832 UP000015103 UP000079169 UP000076502 UP000009046 UP000075886 UP000075840 UP000095301 UP000002320 UP000037069 UP000000673 UP000075885 UP000095300 UP000092444 UP000008820 UP000075884 UP000075881 UP000075880 UP000075900 UP000075901 UP000075902 UP000076407 UP000075882 UP000053105 UP000075920 UP000075903 UP000009192 UP000008792 UP000076408 UP000030765 UP000092443 UP000001070 UP000092445 UP000000304 UP000005408 UP000091820 UP000008711 UP000000803 UP000001292 UP000192221 UP000002282 UP000092460
UP000037510 UP000192223 UP000030742 UP000007266 UP000235965 UP000019118 UP000027135 UP000245037 UP000078542 UP000036403 UP000053825 UP000000311 UP000007755 UP000078540 UP000078541 UP000005205 UP000078492 UP000053097 UP000279307 UP000075809 UP000007819 UP000008237 UP000242457 UP000005203 UP000183832 UP000015103 UP000079169 UP000076502 UP000009046 UP000075886 UP000075840 UP000095301 UP000002320 UP000037069 UP000000673 UP000075885 UP000095300 UP000092444 UP000008820 UP000075884 UP000075881 UP000075880 UP000075900 UP000075901 UP000075902 UP000076407 UP000075882 UP000053105 UP000075920 UP000075903 UP000009192 UP000008792 UP000076408 UP000030765 UP000092443 UP000001070 UP000092445 UP000000304 UP000005408 UP000091820 UP000008711 UP000000803 UP000001292 UP000192221 UP000002282 UP000092460
Interpro
IPR003508
CIDE-N_dom
+ More
IPR015121 DNA_fragmentation_mid_dom
IPR020846 MFS_dom
IPR027417 P-loop_NTPase
IPR003439 ABC_transporter-like
IPR026082 ABCA
IPR003593 AAA+_ATPase
IPR027296 DFF-C
IPR000210 BTB/POZ_dom
IPR006652 Kelch_1
IPR011333 SKP1/BTB/POZ_sf
IPR015915 Kelch-typ_b-propeller
IPR011705 BACK
IPR017299 DFF45
IPR015121 DNA_fragmentation_mid_dom
IPR020846 MFS_dom
IPR027417 P-loop_NTPase
IPR003439 ABC_transporter-like
IPR026082 ABCA
IPR003593 AAA+_ATPase
IPR027296 DFF-C
IPR000210 BTB/POZ_dom
IPR006652 Kelch_1
IPR011333 SKP1/BTB/POZ_sf
IPR015915 Kelch-typ_b-propeller
IPR011705 BACK
IPR017299 DFF45
Gene 3D
CDD
ProteinModelPortal
B0FC96
A0A2H1W0V8
D3JSV7
A0A1E1W5N0
A0A2A4JGA6
A0A194PN41
+ More
S4P8N0 A0A3S2N966 A0A194QMM7 A0A212FJN3 H9JNA5 A0A0L7KP39 A0A1Y1M327 A0A1W4WQE4 J3JZH4 D6WA34 A0A2J7RD86 N6TRH6 A0A067RRT3 A0A2P8Y1C4 A0A2R7W5T4 A0A151IBY9 A0A0J7L3N9 A0A2S2N9B7 A0A0L7QL78 E2A1G4 F4WHK7 A0A151I114 A0A151K1N2 A0A158N900 A0A151J514 A0A2H8TSM0 A0A026VSY1 A0A151WNQ8 E9JAU5 J9JNI3 E2C6N4 A0A2A3ER65 V9IHW8 A0A088AUR4 A0A1J1HMP5 A0A1B6MIX1 A0A1B6GC46 R4G377 A0A1B6DP76 A0A0A9XJA2 A0A1S3CVE3 A0A1Q3EVC5 A0A154PMN3 E0VWD7 A0A182QWK9 A0A182HFQ6 A0A1J1HME3 A0A1I8N5U7 A0A1S4G3C0 B0WG49 A0A0L0C8P1 A0A1J1HRV6 W5J8V0 A0A182P8G5 A0A1I8PYT9 A0A1B0G737 Q16FC1 A0A182MYE3 A0A182JPX1 A0A182J411 A0A182RLP3 A0A182SCB7 A0A182U9F5 A0A182X1U1 A0A182LQ35 A0A0M8ZRN7 A0A182WCU8 A0A182VLQ8 B4KPU8 B4LKV3 A0A182YA41 A0A0A1WNW0 A0A084WI53 A0A034WGY6 A0A023VTR5 A0A0K8U151 A0A1A9XIC9 W8ATX3 A0A224XZA8 B4J5G5 A0A1B0AG69 B4QC70 A0A336MMA2 K1RJK1 A0A1A9WQX9 A0A1S3DKZ7 B3NS92 Q6NR36 B4HNX1 A0A1W4U9Y1 B4P5Q2 A0A1B0BUD3 A0A0B4KFQ3
S4P8N0 A0A3S2N966 A0A194QMM7 A0A212FJN3 H9JNA5 A0A0L7KP39 A0A1Y1M327 A0A1W4WQE4 J3JZH4 D6WA34 A0A2J7RD86 N6TRH6 A0A067RRT3 A0A2P8Y1C4 A0A2R7W5T4 A0A151IBY9 A0A0J7L3N9 A0A2S2N9B7 A0A0L7QL78 E2A1G4 F4WHK7 A0A151I114 A0A151K1N2 A0A158N900 A0A151J514 A0A2H8TSM0 A0A026VSY1 A0A151WNQ8 E9JAU5 J9JNI3 E2C6N4 A0A2A3ER65 V9IHW8 A0A088AUR4 A0A1J1HMP5 A0A1B6MIX1 A0A1B6GC46 R4G377 A0A1B6DP76 A0A0A9XJA2 A0A1S3CVE3 A0A1Q3EVC5 A0A154PMN3 E0VWD7 A0A182QWK9 A0A182HFQ6 A0A1J1HME3 A0A1I8N5U7 A0A1S4G3C0 B0WG49 A0A0L0C8P1 A0A1J1HRV6 W5J8V0 A0A182P8G5 A0A1I8PYT9 A0A1B0G737 Q16FC1 A0A182MYE3 A0A182JPX1 A0A182J411 A0A182RLP3 A0A182SCB7 A0A182U9F5 A0A182X1U1 A0A182LQ35 A0A0M8ZRN7 A0A182WCU8 A0A182VLQ8 B4KPU8 B4LKV3 A0A182YA41 A0A0A1WNW0 A0A084WI53 A0A034WGY6 A0A023VTR5 A0A0K8U151 A0A1A9XIC9 W8ATX3 A0A224XZA8 B4J5G5 A0A1B0AG69 B4QC70 A0A336MMA2 K1RJK1 A0A1A9WQX9 A0A1S3DKZ7 B3NS92 Q6NR36 B4HNX1 A0A1W4U9Y1 B4P5Q2 A0A1B0BUD3 A0A0B4KFQ3
PDB
4D2K
E-value=2.87716e-12,
Score=170
Ontologies
GO
PANTHER
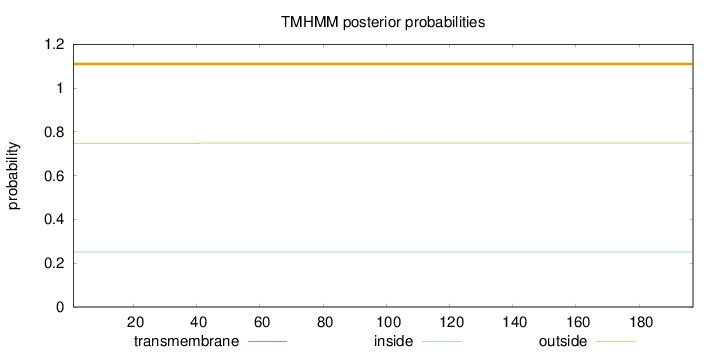
Topology
Length:
197
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00019
Exp number, first 60 AAs:
0.00019
Total prob of N-in:
0.25157
outside
1 - 197
Population Genetic Test Statistics
Pi
293.741321
Theta
225.573463
Tajima's D
0.713408
CLR
11.265281
CSRT
0.574021298935053
Interpretation
Uncertain
