Gene
KWMTBOMO13713 Validated by peptides from experiments
Annotation
prefoldin_subunit_3_[Bombyx_mori]
Full name
Prefoldin subunit 3
Location in the cell
Cytoplasmic Reliability : 2.428 Nuclear Reliability : 2.077
Sequence
CDS
ATGGAAGGAGACGGAGTAGAACCCTCGAACCCTAAGTCGTATTCTGGAATACCAGAAGCTGAATTTGTAGATAATGTAGACGAATTTATGAAATCACCGATCAATGCTGAAGGAGTCGATGTAGTTTTGAAGAGTCTAGATGAAAAGCACCGTAAGTATAAGGTGATGGAATACACGTTGGCTACTAAAAGAAGGCGATTACGGCAACAAATCCCAGACTTAGCGCGGACAATTGAAGTAATTGAAAAATTAAAAGAACAAAAAGAGGAAGTAGAAACACAGTTCCTTCTTAGTGATCAAGTATTCGTGAAGGCTAATGTACCACCAACAAAGTCAGTATGCTTGTGGCTTGGAGCTAATGTAATGTTGGAGTATAGTCTTGAAGATGCTGAAAAATTACTGACTACAAATATGGAAACAGCTCAAGAAAATTTAAATCAAGTAGAACATGATTTGGATTTCTTAAGAGATCAGTGTACCACTACCGAAGTGAATATGGCTAGAGTTTACAACTGGGATGTGAAGAAACGCCAGGCAGCTTCTGGTCGCATTACCACTTGTTAA
Protein
MEGDGVEPSNPKSYSGIPEAEFVDNVDEFMKSPINAEGVDVVLKSLDEKHRKYKVMEYTLATKRRRLRQQIPDLARTIEVIEKLKEQKEEVETQFLLSDQVFVKANVPPTKSVCLWLGANVMLEYSLEDAEKLLTTNMETAQENLNQVEHDLDFLRDQCTTTEVNMARVYNWDVKKRQAASGRITTC
Summary
Description
Binds specifically to cytosolic chaperonin (c-CPN) and transfers target proteins to it. Binds to nascent polypeptide chain and promotes folding in an environment in which there are many competing pathways for nonnative proteins.
Subunit
Heterohexamer of two PFD-alpha type and four PFD-beta type subunits.
Similarity
Belongs to the prefoldin subunit alpha family.
Uniprot
Q1HPR2
A0A1E1W6U4
A0A2H1VZL9
A0A212FJJ2
S4PMW8
A0A194PLE7
+ More
A0A194QN45 A0A0T6B5M5 A0A0L7KN64 A0A1B6HUG4 A0A1B6EQU3 A0A1W4X796 A0A0C9RP12 A0A1B0CZU0 D6WA44 A0A2J7PBH9 A0A2A3EHF6 A0A088AJ58 A0A1B6KKZ5 F4WP05 A0A182X5L9 A0A182HWD0 A0A154NX73 A0A151WM43 A0A0K8TQ52 A0A195E581 A0A310SE73 A5HMP5 A0A195B969 A0A195EUZ8 A0A158NPH9 A0A1S4H5Y6 A0A182KL61 Q7QHF3 A0A0A9WJH4 A0A182UTM6 A0A195CP28 A0A3S2P922 A0A023EHL6 A0A0J7KXA6 T1PGU4 A0A2R7W2Q5 A0A1L8DTZ4 E2ARQ8 J3JZB6 Q17CV4 A0A1Q3F6P6 A0A182RTD7 A0A182NZL6 A0A232FLI8 K7J5F1 A0A182LRI1 A0A034VU72 A0A0A1WTJ0 A0A182JS11 A0A0K8VSE9 A0A161MH09 A0A1Q3F7S9 A0A2M4C0H7 E2BY71 A0A2M4AJ09 W8B403 A0A182QLB8 A0A2M3ZH56 A0A084VHU7 A0A182YB30 A0A182IY41 A0A182N2C3 A0A0L7R7W2 A0A1A9V4P5 A0A1A9XHK7 A0A1B0B480 A0A1A9ZLY0 D3TPL3 A0A1I8PPW1 B0X1A3 T1DJU6 A0A182WDI1 A0A0L0CL29 A0A026W120 T1HC31 T1DNT8 A0A182FKK1 A0A1L8EI82 W5J4H1 A0A1A9WT45 A0A336LIZ2 A0A182T7T2 A0A1Y1NEA9 B3LXC9 B4LXL8 A0A0M4EQ46 U5EVQ5 B3NZF6 E9IST8 Q293S9 B4GLN9 B4NAX9 A0A164X734 V4B4G6
A0A194QN45 A0A0T6B5M5 A0A0L7KN64 A0A1B6HUG4 A0A1B6EQU3 A0A1W4X796 A0A0C9RP12 A0A1B0CZU0 D6WA44 A0A2J7PBH9 A0A2A3EHF6 A0A088AJ58 A0A1B6KKZ5 F4WP05 A0A182X5L9 A0A182HWD0 A0A154NX73 A0A151WM43 A0A0K8TQ52 A0A195E581 A0A310SE73 A5HMP5 A0A195B969 A0A195EUZ8 A0A158NPH9 A0A1S4H5Y6 A0A182KL61 Q7QHF3 A0A0A9WJH4 A0A182UTM6 A0A195CP28 A0A3S2P922 A0A023EHL6 A0A0J7KXA6 T1PGU4 A0A2R7W2Q5 A0A1L8DTZ4 E2ARQ8 J3JZB6 Q17CV4 A0A1Q3F6P6 A0A182RTD7 A0A182NZL6 A0A232FLI8 K7J5F1 A0A182LRI1 A0A034VU72 A0A0A1WTJ0 A0A182JS11 A0A0K8VSE9 A0A161MH09 A0A1Q3F7S9 A0A2M4C0H7 E2BY71 A0A2M4AJ09 W8B403 A0A182QLB8 A0A2M3ZH56 A0A084VHU7 A0A182YB30 A0A182IY41 A0A182N2C3 A0A0L7R7W2 A0A1A9V4P5 A0A1A9XHK7 A0A1B0B480 A0A1A9ZLY0 D3TPL3 A0A1I8PPW1 B0X1A3 T1DJU6 A0A182WDI1 A0A0L0CL29 A0A026W120 T1HC31 T1DNT8 A0A182FKK1 A0A1L8EI82 W5J4H1 A0A1A9WT45 A0A336LIZ2 A0A182T7T2 A0A1Y1NEA9 B3LXC9 B4LXL8 A0A0M4EQ46 U5EVQ5 B3NZF6 E9IST8 Q293S9 B4GLN9 B4NAX9 A0A164X734 V4B4G6
Pubmed
22118469
23622113
26354079
26227816
18362917
19820115
+ More
21719571 26369729 21347285 12364791 20966253 25401762 26823975 24945155 25315136 20798317 22516182 23537049 17510324 28648823 20075255 25348373 25830018 24495485 24438588 25244985 20353571 24330624 26108605 24508170 30249741 20920257 23761445 28004739 17994087 21282665 15632085 23254933
21719571 26369729 21347285 12364791 20966253 25401762 26823975 24945155 25315136 20798317 22516182 23537049 17510324 28648823 20075255 25348373 25830018 24495485 24438588 25244985 20353571 24330624 26108605 24508170 30249741 20920257 23761445 28004739 17994087 21282665 15632085 23254933
EMBL
DQ443340
ABF51429.1
GDQN01009740
GDQN01008379
JAT81314.1
JAT82675.1
+ More
ODYU01005373 SOQ46146.1 AGBW02008253 OWR53917.1 GAIX01003325 JAA89235.1 KQ459600 KPI94152.1 KQ461195 KPJ06779.1 LJIG01009645 KRT82665.1 JTDY01008133 KOB64718.1 GECU01029389 JAS78317.1 GECZ01029482 GECZ01023070 JAS40287.1 JAS46699.1 GBYB01010175 JAG79942.1 AJVK01009723 KQ971312 EEZ98584.1 NEVH01027088 PNF13693.1 KZ288244 PBC31223.1 GEBQ01027858 GEBQ01020031 JAT12119.1 JAT19946.1 GL888243 EGI63936.1 APCN01001485 KQ434777 KZC04162.1 KQ982944 KYQ48888.1 GDAI01001330 JAI16273.1 KQ979608 KYN20345.1 KQ760712 OAD59300.1 EF541362 ABQ18252.1 KQ976542 KYM81081.1 KQ981965 KYN31717.1 ADTU01022553 AAAB01008816 EAA05130.3 GBHO01035022 GBRD01015415 GDHC01012318 JAG08582.1 JAG50411.1 JAQ06311.1 KQ977574 KYN01864.1 RSAL01000184 RVE44846.1 GAPW01004725 JAC08873.1 LBMM01002207 KMQ95142.1 KA647976 AFP62605.1 KK854272 PTY14037.1 GFDF01004161 JAV09923.1 GL442166 EFN63866.1 BT128597 KB631615 AEE63554.1 ERL84675.1 CH477304 EAT44167.1 GFDL01011846 JAV23199.1 NNAY01000065 OXU31369.1 AXCM01000599 GAKP01012081 JAC46871.1 GBXI01011918 JAD02374.1 GDHF01010804 JAI41510.1 GEMB01002656 JAS00536.1 GFDL01011507 JAV23538.1 GGFJ01009669 MBW58810.1 GL451420 EFN79350.1 GGFK01007406 MBW40727.1 GAMC01013263 JAB93292.1 AXCN02001728 GGFM01007017 MBW27768.1 ATLV01013235 KE524847 KFB37541.1 KQ414637 KOC66934.1 JXJN01008196 CCAG010000815 EZ423365 ADD19641.1 DS232256 EDS38552.1 GALA01000051 JAA94801.1 JRES01000238 KNC33063.1 KK107499 QOIP01000003 EZA49770.1 RLU25250.1 ACPB03006253 GAMD01002585 JAA99005.1 GFDG01000434 JAV18365.1 ADMH02002090 ETN59367.1 UFQT01000027 SSX18104.1 GEZM01004809 JAV96304.1 CH902617 EDV42773.1 CH940650 EDW66802.1 CP012526 ALC46372.1 GANO01001771 JAB58100.1 CH954181 EDV49529.1 GL765434 EFZ16415.1 CM000070 EAL29135.1 CH479185 EDW38463.1 CH964232 EDW80943.1 LRGB01001005 KZS13933.1 KB199650 ESP05358.1
ODYU01005373 SOQ46146.1 AGBW02008253 OWR53917.1 GAIX01003325 JAA89235.1 KQ459600 KPI94152.1 KQ461195 KPJ06779.1 LJIG01009645 KRT82665.1 JTDY01008133 KOB64718.1 GECU01029389 JAS78317.1 GECZ01029482 GECZ01023070 JAS40287.1 JAS46699.1 GBYB01010175 JAG79942.1 AJVK01009723 KQ971312 EEZ98584.1 NEVH01027088 PNF13693.1 KZ288244 PBC31223.1 GEBQ01027858 GEBQ01020031 JAT12119.1 JAT19946.1 GL888243 EGI63936.1 APCN01001485 KQ434777 KZC04162.1 KQ982944 KYQ48888.1 GDAI01001330 JAI16273.1 KQ979608 KYN20345.1 KQ760712 OAD59300.1 EF541362 ABQ18252.1 KQ976542 KYM81081.1 KQ981965 KYN31717.1 ADTU01022553 AAAB01008816 EAA05130.3 GBHO01035022 GBRD01015415 GDHC01012318 JAG08582.1 JAG50411.1 JAQ06311.1 KQ977574 KYN01864.1 RSAL01000184 RVE44846.1 GAPW01004725 JAC08873.1 LBMM01002207 KMQ95142.1 KA647976 AFP62605.1 KK854272 PTY14037.1 GFDF01004161 JAV09923.1 GL442166 EFN63866.1 BT128597 KB631615 AEE63554.1 ERL84675.1 CH477304 EAT44167.1 GFDL01011846 JAV23199.1 NNAY01000065 OXU31369.1 AXCM01000599 GAKP01012081 JAC46871.1 GBXI01011918 JAD02374.1 GDHF01010804 JAI41510.1 GEMB01002656 JAS00536.1 GFDL01011507 JAV23538.1 GGFJ01009669 MBW58810.1 GL451420 EFN79350.1 GGFK01007406 MBW40727.1 GAMC01013263 JAB93292.1 AXCN02001728 GGFM01007017 MBW27768.1 ATLV01013235 KE524847 KFB37541.1 KQ414637 KOC66934.1 JXJN01008196 CCAG010000815 EZ423365 ADD19641.1 DS232256 EDS38552.1 GALA01000051 JAA94801.1 JRES01000238 KNC33063.1 KK107499 QOIP01000003 EZA49770.1 RLU25250.1 ACPB03006253 GAMD01002585 JAA99005.1 GFDG01000434 JAV18365.1 ADMH02002090 ETN59367.1 UFQT01000027 SSX18104.1 GEZM01004809 JAV96304.1 CH902617 EDV42773.1 CH940650 EDW66802.1 CP012526 ALC46372.1 GANO01001771 JAB58100.1 CH954181 EDV49529.1 GL765434 EFZ16415.1 CM000070 EAL29135.1 CH479185 EDW38463.1 CH964232 EDW80943.1 LRGB01001005 KZS13933.1 KB199650 ESP05358.1
Proteomes
UP000007151
UP000053268
UP000053240
UP000037510
UP000192223
UP000092462
+ More
UP000007266 UP000235965 UP000242457 UP000005203 UP000007755 UP000076407 UP000075840 UP000076502 UP000075809 UP000078492 UP000078540 UP000078541 UP000005205 UP000075882 UP000007062 UP000075903 UP000078542 UP000283053 UP000036403 UP000095301 UP000000311 UP000030742 UP000008820 UP000075900 UP000075885 UP000215335 UP000002358 UP000075883 UP000075881 UP000008237 UP000075886 UP000030765 UP000076408 UP000075880 UP000075884 UP000053825 UP000078200 UP000092443 UP000092460 UP000092445 UP000092444 UP000095300 UP000002320 UP000075920 UP000037069 UP000053097 UP000279307 UP000015103 UP000069272 UP000000673 UP000091820 UP000075901 UP000007801 UP000008792 UP000092553 UP000008711 UP000001819 UP000008744 UP000007798 UP000076858 UP000030746
UP000007266 UP000235965 UP000242457 UP000005203 UP000007755 UP000076407 UP000075840 UP000076502 UP000075809 UP000078492 UP000078540 UP000078541 UP000005205 UP000075882 UP000007062 UP000075903 UP000078542 UP000283053 UP000036403 UP000095301 UP000000311 UP000030742 UP000008820 UP000075900 UP000075885 UP000215335 UP000002358 UP000075883 UP000075881 UP000008237 UP000075886 UP000030765 UP000076408 UP000075880 UP000075884 UP000053825 UP000078200 UP000092443 UP000092460 UP000092445 UP000092444 UP000095300 UP000002320 UP000075920 UP000037069 UP000053097 UP000279307 UP000015103 UP000069272 UP000000673 UP000091820 UP000075901 UP000007801 UP000008792 UP000092553 UP000008711 UP000001819 UP000008744 UP000007798 UP000076858 UP000030746
PRIDE
Interpro
Gene 3D
ProteinModelPortal
Q1HPR2
A0A1E1W6U4
A0A2H1VZL9
A0A212FJJ2
S4PMW8
A0A194PLE7
+ More
A0A194QN45 A0A0T6B5M5 A0A0L7KN64 A0A1B6HUG4 A0A1B6EQU3 A0A1W4X796 A0A0C9RP12 A0A1B0CZU0 D6WA44 A0A2J7PBH9 A0A2A3EHF6 A0A088AJ58 A0A1B6KKZ5 F4WP05 A0A182X5L9 A0A182HWD0 A0A154NX73 A0A151WM43 A0A0K8TQ52 A0A195E581 A0A310SE73 A5HMP5 A0A195B969 A0A195EUZ8 A0A158NPH9 A0A1S4H5Y6 A0A182KL61 Q7QHF3 A0A0A9WJH4 A0A182UTM6 A0A195CP28 A0A3S2P922 A0A023EHL6 A0A0J7KXA6 T1PGU4 A0A2R7W2Q5 A0A1L8DTZ4 E2ARQ8 J3JZB6 Q17CV4 A0A1Q3F6P6 A0A182RTD7 A0A182NZL6 A0A232FLI8 K7J5F1 A0A182LRI1 A0A034VU72 A0A0A1WTJ0 A0A182JS11 A0A0K8VSE9 A0A161MH09 A0A1Q3F7S9 A0A2M4C0H7 E2BY71 A0A2M4AJ09 W8B403 A0A182QLB8 A0A2M3ZH56 A0A084VHU7 A0A182YB30 A0A182IY41 A0A182N2C3 A0A0L7R7W2 A0A1A9V4P5 A0A1A9XHK7 A0A1B0B480 A0A1A9ZLY0 D3TPL3 A0A1I8PPW1 B0X1A3 T1DJU6 A0A182WDI1 A0A0L0CL29 A0A026W120 T1HC31 T1DNT8 A0A182FKK1 A0A1L8EI82 W5J4H1 A0A1A9WT45 A0A336LIZ2 A0A182T7T2 A0A1Y1NEA9 B3LXC9 B4LXL8 A0A0M4EQ46 U5EVQ5 B3NZF6 E9IST8 Q293S9 B4GLN9 B4NAX9 A0A164X734 V4B4G6
A0A194QN45 A0A0T6B5M5 A0A0L7KN64 A0A1B6HUG4 A0A1B6EQU3 A0A1W4X796 A0A0C9RP12 A0A1B0CZU0 D6WA44 A0A2J7PBH9 A0A2A3EHF6 A0A088AJ58 A0A1B6KKZ5 F4WP05 A0A182X5L9 A0A182HWD0 A0A154NX73 A0A151WM43 A0A0K8TQ52 A0A195E581 A0A310SE73 A5HMP5 A0A195B969 A0A195EUZ8 A0A158NPH9 A0A1S4H5Y6 A0A182KL61 Q7QHF3 A0A0A9WJH4 A0A182UTM6 A0A195CP28 A0A3S2P922 A0A023EHL6 A0A0J7KXA6 T1PGU4 A0A2R7W2Q5 A0A1L8DTZ4 E2ARQ8 J3JZB6 Q17CV4 A0A1Q3F6P6 A0A182RTD7 A0A182NZL6 A0A232FLI8 K7J5F1 A0A182LRI1 A0A034VU72 A0A0A1WTJ0 A0A182JS11 A0A0K8VSE9 A0A161MH09 A0A1Q3F7S9 A0A2M4C0H7 E2BY71 A0A2M4AJ09 W8B403 A0A182QLB8 A0A2M3ZH56 A0A084VHU7 A0A182YB30 A0A182IY41 A0A182N2C3 A0A0L7R7W2 A0A1A9V4P5 A0A1A9XHK7 A0A1B0B480 A0A1A9ZLY0 D3TPL3 A0A1I8PPW1 B0X1A3 T1DJU6 A0A182WDI1 A0A0L0CL29 A0A026W120 T1HC31 T1DNT8 A0A182FKK1 A0A1L8EI82 W5J4H1 A0A1A9WT45 A0A336LIZ2 A0A182T7T2 A0A1Y1NEA9 B3LXC9 B4LXL8 A0A0M4EQ46 U5EVQ5 B3NZF6 E9IST8 Q293S9 B4GLN9 B4NAX9 A0A164X734 V4B4G6
Ontologies
GO
PANTHER
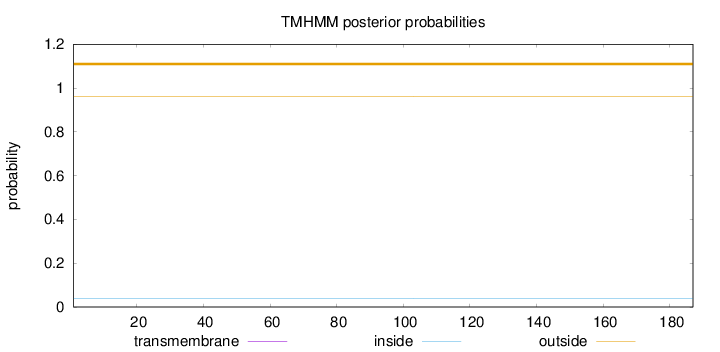
Topology
Length:
187
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0012
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03996
outside
1 - 187
Population Genetic Test Statistics
Pi
193.71185
Theta
153.455529
Tajima's D
0.408877
CLR
0.541013
CSRT
0.486325683715814
Interpretation
Uncertain
