Gene
KWMTBOMO13711
Pre Gene Modal
BGIBMGA011011
Annotation
PREDICTED:_monocarboxylate_transporter_10_[Papilio_polytes]
Location in the cell
PlasmaMembrane Reliability : 4.978
Sequence
CDS
ATGAACCTCGACCCAAAAGAAAATTCAGACTTACTTCAAGAAAGGAGCAAGGTAGATACTAACGGTAATGCTAATCAGGAAGAAGTCCGAGAGCCTCCTGATGGTGGTTTCCGAGCTTACGCGGTTGTTCTCGCATCATTTTTTACCAACGGACTGCTTTTTGGGGTAATAAATTCCTGCAGTGTTGTATATCCCGTTCTCGAGATAATATTAAAAGAACAGGGAGCCACAAATGAAGAGAGCCGTGCTGCTCTGGTTGGTGCCTTGACTATGGGTATGACGTTTTTGTTGTCGCCTTTGTCTGGAGTGCTGACAGGAATAGTGGGAATACGCTTGACGGCTGTATTAGGCGGAACCATCGCCGCACTTGGTTTAATTGTATCTTCGTTCGTTGTTGACAACATCAATGCTCTCTATTTTACATATAGTATTATGTATGGTCTGGGAGCATCTTTTGCTTATACCCCGTCTCTAGCAATATTGGGACACTATTTCAAAAAACACTTGGGCTTAGTTAACGGTCTTGTAACAGTAGGAAGCTCTGTTTTCACTGCGTTCATACCTTCCCTCATGGAGTATATGCTGAATAATTACGGTTTGCAGTGGTTGTTTCGGTTCTTAGGTCTTTTGACATTCGGCGTTGCTCTTTGCGGCTTGCTGTTTAAGCCCTTAATGAATATTCCAAAACCTTCGAGAAGAGATGGAAATCTCAAATCCTTTTTAACGAAAATAATCAATATACAAATTTGGAAAAACAGAAAGTATAGATTTTGGGCATTATCGATGCCTGTAGCTTTGTTTGGCTATTTTGTGAGTTACGTTCACATTAAAAAGTTTATGAATAATAATTTTCCCGGTGCTAATTACAATGTGCCTTTACAGTGCATAGCGATAACATCTGGTATGGGTAGATTATTGTTTGGAGTTTTAGCTGATAAACCAGGTATAAACAAAATTTTGTTGCAACAAATTTCTTTTTATGTAATAGGAATTCTGACAATAGCTTTGCCATTTGTTAGATCTTTCTCCATGGTAGTGGTCATTTCACTTGGAATGGGTATATTTGATGGAGCGTTCATCGCCCTCATAGGGCCAATCGCTATTCAGTTATGTGGTAGAGCATACGCAGCTCAAGCAATAGGTTGCATGTTAGGGATGTCTGCAATGCCACTTTCCGCGGGTCCACCGATCGCTGCTTATATACATCGATTAAGTGGATCTTATACGTTACCTTTCGTTTTGGCGGGAATATCGCCTATTGTTGGGGCAACTTTAATGTTTTCCGTACATTTCCAACGGCAAAATGGAGAGACTGAAATTGTTACTAATGGCCACGCTCCTTCAGATGTTGAAAAATCGATACCGCTGCTAGTTTCAAATGGGAATGCTAAACAACAGACAGCTTTATAA
Protein
MNLDPKENSDLLQERSKVDTNGNANQEEVREPPDGGFRAYAVVLASFFTNGLLFGVINSCSVVYPVLEIILKEQGATNEESRAALVGALTMGMTFLLSPLSGVLTGIVGIRLTAVLGGTIAALGLIVSSFVVDNINALYFTYSIMYGLGASFAYTPSLAILGHYFKKHLGLVNGLVTVGSSVFTAFIPSLMEYMLNNYGLQWLFRFLGLLTFGVALCGLLFKPLMNIPKPSRRDGNLKSFLTKIINIQIWKNRKYRFWALSMPVALFGYFVSYVHIKKFMNNNFPGANYNVPLQCIAITSGMGRLLFGVLADKPGINKILLQQISFYVIGILTIALPFVRSFSMVVVISLGMGIFDGAFIALIGPIAIQLCGRAYAAQAIGCMLGMSAMPLSAGPPIAAYIHRLSGSYTLPFVLAGISPIVGATLMFSVHFQRQNGETEIVTNGHAPSDVEKSIPLLVSNGNAKQQTAL
Summary
Uniprot
H9JNA8
A0A2H1VZC8
A0A194QTB1
A0A2A4J4T8
A0A212FJK4
A0A3S2LUV9
+ More
A0A0L7L0E4 A0A2A4J4V8 C6KEM2 A0A0L7RF68 E2ALF2 A0A151J430 A0A195FSD5 A0A158NSL0 A0A182H4B9 A0A151HZ27 F4WFK3 A0A087ZXB5 A0A151WI84 A0A1Q3EUJ5 A0A195CSS6 A0A232F8R6 A0A1A9ZXQ5 E2BW41 A0A1S4GW22 Q7Q374 A0A182V1I6 A0A1B0B354 A0A182TE48 A0A3L8DX54 D3TPV6 A0A1L8DEH1 A0A2A3E4N9 B4K766 A0A0Q9X206 A0A1A9VEU1 A0A1A9XZ83 A0A0A1XG97 B4JRS8 A0A1B0C9L0 T1E1W7 A0A0A1X1Z9 A0A0Q9X926 A0A0M4F3Y7 A0A2M3ZGE1 A0A2M3ZGD0 A0A2M3ZGJ8 A0A1A9X1I8 A0A084WHE6 A0A310SGW3 A0A182JQL9 A0A336LEE7 A0A2M4CQ03 A0A182PCX4 A0A2M4CPZ5 A0A0K8U3P0 A0A182XZS4 A0A2M4BKY8 A0A2M4BL06 A0A0K8VD47 A0A2M4A083 A0A034V2X8 A0A182F669 A0A2M4A054 A0A182MVS5 A0A182SRY1 A0A026WDA8 U5ESF8 A0A0C9R454 A0A336MQP3 A0A336L5I9 A0A0Q9WSS6 B4M096 A0A0L0C0Y0 Q299V0 W8B3K1 W8BEK1 B4G5S2 A0A1B6D6R1 A0A3B0JDB9 A0A0R3NH36 A0A1B0TRG3 B4NI09 D6WAV9 A0A067RRU3 A0A1Q3EUU9 A0A1W4UAW4 A0A1I8Q065 A0A1B6GSW9 A0A1J1ICA4 A0A1I8Q0A0 Q95SN6 A0A1W4UPR6 A0A1I8MLJ7 T1PGG4 B4PPD5 B4HGU2 A0A0J7KKR5 Q9VG39
A0A0L7L0E4 A0A2A4J4V8 C6KEM2 A0A0L7RF68 E2ALF2 A0A151J430 A0A195FSD5 A0A158NSL0 A0A182H4B9 A0A151HZ27 F4WFK3 A0A087ZXB5 A0A151WI84 A0A1Q3EUJ5 A0A195CSS6 A0A232F8R6 A0A1A9ZXQ5 E2BW41 A0A1S4GW22 Q7Q374 A0A182V1I6 A0A1B0B354 A0A182TE48 A0A3L8DX54 D3TPV6 A0A1L8DEH1 A0A2A3E4N9 B4K766 A0A0Q9X206 A0A1A9VEU1 A0A1A9XZ83 A0A0A1XG97 B4JRS8 A0A1B0C9L0 T1E1W7 A0A0A1X1Z9 A0A0Q9X926 A0A0M4F3Y7 A0A2M3ZGE1 A0A2M3ZGD0 A0A2M3ZGJ8 A0A1A9X1I8 A0A084WHE6 A0A310SGW3 A0A182JQL9 A0A336LEE7 A0A2M4CQ03 A0A182PCX4 A0A2M4CPZ5 A0A0K8U3P0 A0A182XZS4 A0A2M4BKY8 A0A2M4BL06 A0A0K8VD47 A0A2M4A083 A0A034V2X8 A0A182F669 A0A2M4A054 A0A182MVS5 A0A182SRY1 A0A026WDA8 U5ESF8 A0A0C9R454 A0A336MQP3 A0A336L5I9 A0A0Q9WSS6 B4M096 A0A0L0C0Y0 Q299V0 W8B3K1 W8BEK1 B4G5S2 A0A1B6D6R1 A0A3B0JDB9 A0A0R3NH36 A0A1B0TRG3 B4NI09 D6WAV9 A0A067RRU3 A0A1Q3EUU9 A0A1W4UAW4 A0A1I8Q065 A0A1B6GSW9 A0A1J1ICA4 A0A1I8Q0A0 Q95SN6 A0A1W4UPR6 A0A1I8MLJ7 T1PGG4 B4PPD5 B4HGU2 A0A0J7KKR5 Q9VG39
Pubmed
19121390
26354079
22118469
26227816
19754707
20798317
+ More
21347285 26483478 21719571 28648823 12364791 30249741 20353571 17994087 25830018 24330624 18057021 24438588 25244985 25348373 24508170 26108605 15632085 24495485 18362917 19820115 24845553 25315136 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
21347285 26483478 21719571 28648823 12364791 30249741 20353571 17994087 25830018 24330624 18057021 24438588 25244985 25348373 24508170 26108605 15632085 24495485 18362917 19820115 24845553 25315136 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
BABH01019626
ODYU01005373
SOQ46147.1
KQ461195
KPJ06781.1
NWSH01003372
+ More
PCG66433.1 AGBW02008253 OWR53916.1 RSAL01000184 RVE44855.1 JTDY01003954 KOB68781.1 PCG66432.1 GQ184571 ACS75760.1 KQ414605 KOC69627.1 GL440604 EFN65757.1 KQ980211 KYN17202.1 KQ981276 KYN43500.1 ADTU01024945 ADTU01024946 JXUM01001208 JXUM01001209 JXUM01001210 JXUM01001211 JXUM01001212 KQ976719 KYM76742.1 GL888120 EGI66980.1 KQ983089 KYQ47579.1 GFDL01016068 JAV18977.1 KQ977329 KYN03577.1 NNAY01000681 OXU27002.1 GL451091 EFN80073.1 AAAB01008964 EAA12383.3 JXJN01007770 QOIP01000003 RLU24368.1 EZ423458 ADD19734.1 GFDF01009221 JAV04863.1 KZ288376 PBC26658.1 CH933806 EDW16379.2 KRG02075.1 GBXI01010244 GBXI01004321 JAD04048.1 JAD09971.1 CH916373 EDV94468.1 AJWK01002452 AJWK01002453 GALA01001705 JAA93147.1 GBXI01008953 JAD05339.1 KRG02074.1 KRG02076.1 KRG02077.1 CP012526 ALC46387.1 GGFM01006873 MBW27624.1 GGFM01006855 MBW27606.1 GGFM01006854 MBW27605.1 ATLV01023808 KE525346 KFB49640.1 KQ760123 OAD61860.1 UFQS01001808 UFQT01001808 SSX12402.1 SSX31853.1 GGFL01003232 MBW67410.1 GGFL01003209 MBW67387.1 GDHF01031136 JAI21178.1 GGFJ01004595 MBW53736.1 GGFJ01004594 MBW53735.1 GDHF01015521 JAI36793.1 GGFK01000834 MBW34155.1 GAKP01022455 GAKP01022454 JAC36497.1 GGFK01000801 MBW34122.1 AXCM01001390 KK107295 EZA53034.1 GANO01003265 JAB56606.1 GBYB01001595 GBYB01001596 JAG71362.1 JAG71363.1 SSX12400.1 SSX31851.1 SSX12401.1 SSX31852.1 CH940650 KRF83216.1 EDW67258.1 KRF83215.1 KRF83217.1 KRF83218.1 JRES01001061 KNC25917.1 CM000070 EAL27601.2 GAMC01014922 JAB91633.1 GAMC01014924 GAMC01014923 JAB91631.1 CH479179 EDW24938.1 GEDC01015952 JAS21346.1 OUUW01000005 SPP80374.1 KRT00172.1 KT304312 ALQ52683.1 CH964272 EDW83659.2 KQ971312 EEZ98692.2 KK852463 KDR23400.1 GFDL01016067 JAV18978.1 GECZ01004241 JAS65528.1 CVRI01000047 CRK97917.1 AY060688 AAL28236.1 KA647230 AFP61859.1 CM000160 EDW97141.1 CH480815 EDW42413.1 LBMM01006205 KMQ90811.1 AE014297 BT021229 AAF54851.2 AAX33377.1
PCG66433.1 AGBW02008253 OWR53916.1 RSAL01000184 RVE44855.1 JTDY01003954 KOB68781.1 PCG66432.1 GQ184571 ACS75760.1 KQ414605 KOC69627.1 GL440604 EFN65757.1 KQ980211 KYN17202.1 KQ981276 KYN43500.1 ADTU01024945 ADTU01024946 JXUM01001208 JXUM01001209 JXUM01001210 JXUM01001211 JXUM01001212 KQ976719 KYM76742.1 GL888120 EGI66980.1 KQ983089 KYQ47579.1 GFDL01016068 JAV18977.1 KQ977329 KYN03577.1 NNAY01000681 OXU27002.1 GL451091 EFN80073.1 AAAB01008964 EAA12383.3 JXJN01007770 QOIP01000003 RLU24368.1 EZ423458 ADD19734.1 GFDF01009221 JAV04863.1 KZ288376 PBC26658.1 CH933806 EDW16379.2 KRG02075.1 GBXI01010244 GBXI01004321 JAD04048.1 JAD09971.1 CH916373 EDV94468.1 AJWK01002452 AJWK01002453 GALA01001705 JAA93147.1 GBXI01008953 JAD05339.1 KRG02074.1 KRG02076.1 KRG02077.1 CP012526 ALC46387.1 GGFM01006873 MBW27624.1 GGFM01006855 MBW27606.1 GGFM01006854 MBW27605.1 ATLV01023808 KE525346 KFB49640.1 KQ760123 OAD61860.1 UFQS01001808 UFQT01001808 SSX12402.1 SSX31853.1 GGFL01003232 MBW67410.1 GGFL01003209 MBW67387.1 GDHF01031136 JAI21178.1 GGFJ01004595 MBW53736.1 GGFJ01004594 MBW53735.1 GDHF01015521 JAI36793.1 GGFK01000834 MBW34155.1 GAKP01022455 GAKP01022454 JAC36497.1 GGFK01000801 MBW34122.1 AXCM01001390 KK107295 EZA53034.1 GANO01003265 JAB56606.1 GBYB01001595 GBYB01001596 JAG71362.1 JAG71363.1 SSX12400.1 SSX31851.1 SSX12401.1 SSX31852.1 CH940650 KRF83216.1 EDW67258.1 KRF83215.1 KRF83217.1 KRF83218.1 JRES01001061 KNC25917.1 CM000070 EAL27601.2 GAMC01014922 JAB91633.1 GAMC01014924 GAMC01014923 JAB91631.1 CH479179 EDW24938.1 GEDC01015952 JAS21346.1 OUUW01000005 SPP80374.1 KRT00172.1 KT304312 ALQ52683.1 CH964272 EDW83659.2 KQ971312 EEZ98692.2 KK852463 KDR23400.1 GFDL01016067 JAV18978.1 GECZ01004241 JAS65528.1 CVRI01000047 CRK97917.1 AY060688 AAL28236.1 KA647230 AFP61859.1 CM000160 EDW97141.1 CH480815 EDW42413.1 LBMM01006205 KMQ90811.1 AE014297 BT021229 AAF54851.2 AAX33377.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000007151
UP000283053
UP000037510
+ More
UP000053825 UP000000311 UP000078492 UP000078541 UP000005205 UP000069940 UP000078540 UP000007755 UP000005203 UP000075809 UP000078542 UP000215335 UP000092445 UP000008237 UP000007062 UP000075903 UP000092460 UP000075902 UP000279307 UP000242457 UP000009192 UP000078200 UP000092443 UP000001070 UP000092461 UP000092553 UP000091820 UP000030765 UP000075881 UP000075885 UP000076408 UP000069272 UP000075883 UP000075901 UP000053097 UP000008792 UP000037069 UP000001819 UP000008744 UP000268350 UP000007798 UP000007266 UP000027135 UP000192221 UP000095300 UP000183832 UP000095301 UP000002282 UP000001292 UP000036403 UP000000803
UP000053825 UP000000311 UP000078492 UP000078541 UP000005205 UP000069940 UP000078540 UP000007755 UP000005203 UP000075809 UP000078542 UP000215335 UP000092445 UP000008237 UP000007062 UP000075903 UP000092460 UP000075902 UP000279307 UP000242457 UP000009192 UP000078200 UP000092443 UP000001070 UP000092461 UP000092553 UP000091820 UP000030765 UP000075881 UP000075885 UP000076408 UP000069272 UP000075883 UP000075901 UP000053097 UP000008792 UP000037069 UP000001819 UP000008744 UP000268350 UP000007798 UP000007266 UP000027135 UP000192221 UP000095300 UP000183832 UP000095301 UP000002282 UP000001292 UP000036403 UP000000803
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JNA8
A0A2H1VZC8
A0A194QTB1
A0A2A4J4T8
A0A212FJK4
A0A3S2LUV9
+ More
A0A0L7L0E4 A0A2A4J4V8 C6KEM2 A0A0L7RF68 E2ALF2 A0A151J430 A0A195FSD5 A0A158NSL0 A0A182H4B9 A0A151HZ27 F4WFK3 A0A087ZXB5 A0A151WI84 A0A1Q3EUJ5 A0A195CSS6 A0A232F8R6 A0A1A9ZXQ5 E2BW41 A0A1S4GW22 Q7Q374 A0A182V1I6 A0A1B0B354 A0A182TE48 A0A3L8DX54 D3TPV6 A0A1L8DEH1 A0A2A3E4N9 B4K766 A0A0Q9X206 A0A1A9VEU1 A0A1A9XZ83 A0A0A1XG97 B4JRS8 A0A1B0C9L0 T1E1W7 A0A0A1X1Z9 A0A0Q9X926 A0A0M4F3Y7 A0A2M3ZGE1 A0A2M3ZGD0 A0A2M3ZGJ8 A0A1A9X1I8 A0A084WHE6 A0A310SGW3 A0A182JQL9 A0A336LEE7 A0A2M4CQ03 A0A182PCX4 A0A2M4CPZ5 A0A0K8U3P0 A0A182XZS4 A0A2M4BKY8 A0A2M4BL06 A0A0K8VD47 A0A2M4A083 A0A034V2X8 A0A182F669 A0A2M4A054 A0A182MVS5 A0A182SRY1 A0A026WDA8 U5ESF8 A0A0C9R454 A0A336MQP3 A0A336L5I9 A0A0Q9WSS6 B4M096 A0A0L0C0Y0 Q299V0 W8B3K1 W8BEK1 B4G5S2 A0A1B6D6R1 A0A3B0JDB9 A0A0R3NH36 A0A1B0TRG3 B4NI09 D6WAV9 A0A067RRU3 A0A1Q3EUU9 A0A1W4UAW4 A0A1I8Q065 A0A1B6GSW9 A0A1J1ICA4 A0A1I8Q0A0 Q95SN6 A0A1W4UPR6 A0A1I8MLJ7 T1PGG4 B4PPD5 B4HGU2 A0A0J7KKR5 Q9VG39
A0A0L7L0E4 A0A2A4J4V8 C6KEM2 A0A0L7RF68 E2ALF2 A0A151J430 A0A195FSD5 A0A158NSL0 A0A182H4B9 A0A151HZ27 F4WFK3 A0A087ZXB5 A0A151WI84 A0A1Q3EUJ5 A0A195CSS6 A0A232F8R6 A0A1A9ZXQ5 E2BW41 A0A1S4GW22 Q7Q374 A0A182V1I6 A0A1B0B354 A0A182TE48 A0A3L8DX54 D3TPV6 A0A1L8DEH1 A0A2A3E4N9 B4K766 A0A0Q9X206 A0A1A9VEU1 A0A1A9XZ83 A0A0A1XG97 B4JRS8 A0A1B0C9L0 T1E1W7 A0A0A1X1Z9 A0A0Q9X926 A0A0M4F3Y7 A0A2M3ZGE1 A0A2M3ZGD0 A0A2M3ZGJ8 A0A1A9X1I8 A0A084WHE6 A0A310SGW3 A0A182JQL9 A0A336LEE7 A0A2M4CQ03 A0A182PCX4 A0A2M4CPZ5 A0A0K8U3P0 A0A182XZS4 A0A2M4BKY8 A0A2M4BL06 A0A0K8VD47 A0A2M4A083 A0A034V2X8 A0A182F669 A0A2M4A054 A0A182MVS5 A0A182SRY1 A0A026WDA8 U5ESF8 A0A0C9R454 A0A336MQP3 A0A336L5I9 A0A0Q9WSS6 B4M096 A0A0L0C0Y0 Q299V0 W8B3K1 W8BEK1 B4G5S2 A0A1B6D6R1 A0A3B0JDB9 A0A0R3NH36 A0A1B0TRG3 B4NI09 D6WAV9 A0A067RRU3 A0A1Q3EUU9 A0A1W4UAW4 A0A1I8Q065 A0A1B6GSW9 A0A1J1ICA4 A0A1I8Q0A0 Q95SN6 A0A1W4UPR6 A0A1I8MLJ7 T1PGG4 B4PPD5 B4HGU2 A0A0J7KKR5 Q9VG39
Ontologies
GO
PANTHER
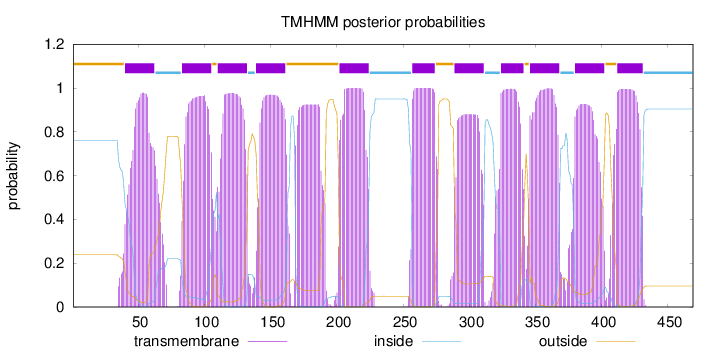
Topology
Length:
469
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
252.20709
Exp number, first 60 AAs:
17.36872
Total prob of N-in:
0.76181
POSSIBLE N-term signal
sequence
outside
1 - 39
TMhelix
40 - 62
inside
63 - 82
TMhelix
83 - 105
outside
106 - 109
TMhelix
110 - 132
inside
133 - 138
TMhelix
139 - 161
outside
162 - 201
TMhelix
202 - 224
inside
225 - 256
TMhelix
257 - 274
outside
275 - 288
TMhelix
289 - 311
inside
312 - 323
TMhelix
324 - 341
outside
342 - 345
TMhelix
346 - 368
inside
369 - 379
TMhelix
380 - 402
outside
403 - 411
TMhelix
412 - 431
inside
432 - 469
Population Genetic Test Statistics
Pi
218.080821
Theta
163.437755
Tajima's D
1.112429
CLR
0
CSRT
0.693815309234538
Interpretation
Possibly Positive selection
