Pre Gene Modal
BGIBMGA011012
Annotation
PREDICTED:_organic_cation_transporter_protein_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.851
Sequence
CDS
ATGACGGCTGGTATGCATATGCTGTCTTTAGTGACAGTGGCCGCAGTTCCGGATCACAGATGCTTCCTACACGGAATTGACAGCTACAATGCCACAAATTCATGGAACTCTTCTTTGGTAATGGCAGCTATTCCAGTGAATGAAAATGACAAATTTGATTCGTGCAGAATGTACACAAGGAACCACACATCTACGGCGGAATGTGAATCTTGGGTTTACAACACGCAGTACTTTTCTTCTTCTAGGGGTATTGAATGGAATTTTGTATGCAATCAGCGTTGGATGGGCGCTATCGCTCAAACCGTTTACATGTTTGGCGTTGTGGGTGGCTCTCTCATATTTGGTCGAATATCTGATAAATATGGAAGGAAAACTGCCTTTGTATGGTCTGGAGTTATGCAATTGCTGCTCGGAGTTTTGGTTGCATTAACAAATGAATACTACTCTTTCTTAGTTGTCAGATTTATATATGGAATATTTGGATCGGGAGCATATATTACTGGATTCGTATTAACGATGGAACTCGTCGGACCAAGCAAACGAACAATTTGCGGGGTTACATTTCAAGTAATGTTCGCAATCGGGATTATGTTAGTTCCTGCTTGGGGATATTTAATTGATAATAGATTTTACTTGCAAATACTGTATGGTCTGCACGCAGCTCTGTTACTTCCGCACTGGTTTATAATGGACGAGTCCCCAAGGTGGCTTTGGTCACAAGGCCGTGCTCGAGAATCGGTCGCAATAATTGAAAAAGCGTTGAAGATGAACGGTATAACTGAGAACATAGATACGTCTTCTTTAATTTCACAATGTAAAGCGACAAATGCAAAATATTCAGATGAACAATCTGCGGGCACGAATGATCTTTTCAAGACTCCAAGAATGTTGAAGAAAACTCTTATAATTTGCGGATGCTGGTTCACAAACTCCGTTGTCTATTATGGGATTTCATTGAACGCTGGAAAATTAAATGGAAACCCATATTTTCTTATGTTCTTAATGGGTATCGTAGAGTTACCTAGCTATGTAATTATAACTTACTTCTTGGACCGGGTTGGACACAGAGCTCTTATTAGTACAATGATGTTGCTAGGTGGAATAGCATGTTTGGTTATATTCTTTCTTCCTCATGGATCTATAACTACCACTGGAATGGTAATGGTAGGAAAGCTTTTCATTTCAGGTTCATTCTCTATTATTTACAAGTATTCTGCAGAATTATTCCCGACCGTTGTTCGTAGTTCTGGAGTTGGTCTGGGCAGTATGTGCGCCAGTGTGTCCGGAGCTTTAACCCCCCTGATAAATTTATTAGATACCCTAAATCCCAAGATTCCGACTATTTTATTCGGATTTTTGGCTTTATTCTCTGGGTTCTCTACTTTCTTCTTACCGGAAACCATTGGCCGCCACTTACCCCAATCTCTGGAAGATGGAGAAAAATTCGGAATCGGCGACAATTGTTTCACCAACTGCAGTGGTCAGCCACTGACTGATAGTATTCCCGATATTCCTGAAACCATGGTTCCTCTAGATGAAAACAAGAAAACAAAGGAATAA
Protein
MTAGMHMLSLVTVAAVPDHRCFLHGIDSYNATNSWNSSLVMAAIPVNENDKFDSCRMYTRNHTSTAECESWVYNTQYFSSSRGIEWNFVCNQRWMGAIAQTVYMFGVVGGSLIFGRISDKYGRKTAFVWSGVMQLLLGVLVALTNEYYSFLVVRFIYGIFGSGAYITGFVLTMELVGPSKRTICGVTFQVMFAIGIMLVPAWGYLIDNRFYLQILYGLHAALLLPHWFIMDESPRWLWSQGRARESVAIIEKALKMNGITENIDTSSLISQCKATNAKYSDEQSAGTNDLFKTPRMLKKTLIICGCWFTNSVVYYGISLNAGKLNGNPYFLMFLMGIVELPSYVIITYFLDRVGHRALISTMMLLGGIACLVIFFLPHGSITTTGMVMVGKLFISGSFSIIYKYSAELFPTVVRSSGVGLGSMCASVSGALTPLINLLDTLNPKIPTILFGFLALFSGFSTFFLPETIGRHLPQSLEDGEKFGIGDNCFTNCSGQPLTDSIPDIPETMVPLDENKKTKE
Summary
Uniprot
H9JNA9
A0A2H1VCG7
A0A0L7LRI0
A0A194PLE2
A0A194QMN2
A0A212FJK2
+ More
A0A2A4J2Z5 A0A1I8M620 T1PFX4 A0A084WAL6 A0A182RIT6 A0A1I8PLQ6 A0A1L8EFV0 A0A182VJP9 A0A182HYF0 A0A182LPX9 A0A182WUV5 A0A182J953 A0A182UC81 A0A182YF87 B3P0Z4 A0A182QVY2 Q7Q8X0 B4NKZ9 A0A182VT64 Q294V9 B4PR27 B3MTU9 A0A182K4T8 A0A2M4ACZ2 B4HLC5 A0A2M4ABC8 Q961R9 A0A182FTH7 T1DSK2 A0A0M3QX72 A0A2M4CUC2 A0A2M3Z135 A0A2M4BJB2 A0A182MLM8 A0A2M3Z0U5 A0A2M4CUK7 A0A2M3ZKS7 B0X0R5 A0A0L0BNY0 B4KA02 A0A182NDC5 A0A3B0KF16 A0A1W4VFN7 A0A2H1VCJ0 A0A0T6B785 B4QX65 U5EUX6 A0A1Q3FFR9 A0A1Q3FBY2 A0A1Q3FFU4 A0A1Q3FC36 A0A023ETV6 A0A182PJ93 B4LXZ8 A0A3B0JTC4 Q17MW7 H9JNB0 B4JG18 A0A1B0C9K9 A0A1S4H482 A0A1B0GCT6 A0A1A9ZM58 A0A034WJ19 A0A1B0D350 A0A0K8UQD0 A0A0A1X2V9 W8AV18 A0A1L8DER9 A0A1L8DEC5 A0A1A9VAZ8 A0A1A9WHP3 A0A194QMM0 D6WAW4 A0A154PMR9 A0A1B6KJY1 A0A1A9XU90 A0A1B0BNC8 A0A1B6H694 A0A1B6G772 A0A1J1IED5 A0A310SKE6 A0A0K8TNM3 A0A026VYB7 A0A067RHW4 A0A2A3EJV7 A0A1W4XL70 A0A087ZUB5 E9JC16 N6TSC1 A0A0L7QWS4 A0A1W4XLF5 E2BB78 A0A1Y1JWN7 A0A1Y1JSF9
A0A2A4J2Z5 A0A1I8M620 T1PFX4 A0A084WAL6 A0A182RIT6 A0A1I8PLQ6 A0A1L8EFV0 A0A182VJP9 A0A182HYF0 A0A182LPX9 A0A182WUV5 A0A182J953 A0A182UC81 A0A182YF87 B3P0Z4 A0A182QVY2 Q7Q8X0 B4NKZ9 A0A182VT64 Q294V9 B4PR27 B3MTU9 A0A182K4T8 A0A2M4ACZ2 B4HLC5 A0A2M4ABC8 Q961R9 A0A182FTH7 T1DSK2 A0A0M3QX72 A0A2M4CUC2 A0A2M3Z135 A0A2M4BJB2 A0A182MLM8 A0A2M3Z0U5 A0A2M4CUK7 A0A2M3ZKS7 B0X0R5 A0A0L0BNY0 B4KA02 A0A182NDC5 A0A3B0KF16 A0A1W4VFN7 A0A2H1VCJ0 A0A0T6B785 B4QX65 U5EUX6 A0A1Q3FFR9 A0A1Q3FBY2 A0A1Q3FFU4 A0A1Q3FC36 A0A023ETV6 A0A182PJ93 B4LXZ8 A0A3B0JTC4 Q17MW7 H9JNB0 B4JG18 A0A1B0C9K9 A0A1S4H482 A0A1B0GCT6 A0A1A9ZM58 A0A034WJ19 A0A1B0D350 A0A0K8UQD0 A0A0A1X2V9 W8AV18 A0A1L8DER9 A0A1L8DEC5 A0A1A9VAZ8 A0A1A9WHP3 A0A194QMM0 D6WAW4 A0A154PMR9 A0A1B6KJY1 A0A1A9XU90 A0A1B0BNC8 A0A1B6H694 A0A1B6G772 A0A1J1IED5 A0A310SKE6 A0A0K8TNM3 A0A026VYB7 A0A067RHW4 A0A2A3EJV7 A0A1W4XL70 A0A087ZUB5 E9JC16 N6TSC1 A0A0L7QWS4 A0A1W4XLF5 E2BB78 A0A1Y1JWN7 A0A1Y1JSF9
Pubmed
19121390
26227816
26354079
22118469
25315136
24438588
+ More
20966253 25244985 17994087 12364791 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 24945155 17510324 25348373 25830018 24495485 18362917 19820115 26369729 24508170 30249741 24845553 21282665 23537049 20798317 28004739
20966253 25244985 17994087 12364791 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605 24945155 17510324 25348373 25830018 24495485 18362917 19820115 26369729 24508170 30249741 24845553 21282665 23537049 20798317 28004739
EMBL
BABH01019619
BABH01019620
ODYU01001804
SOQ38505.1
JTDY01000250
KOB78065.1
+ More
KQ459600 KPI94147.1 KQ461195 KPJ06782.1 AGBW02008253 OWR53915.1 NWSH01003372 PCG66435.1 KA647574 AFP62203.1 ATLV01022206 KE525330 KFB47260.1 GFDG01001231 JAV17568.1 APCN01005901 CH954181 EDV49113.1 AXCN02001770 AAAB01008933 EAA09977.4 CH964272 EDW84202.1 CM000070 EAL28854.1 CM000160 EDW97349.1 CH902623 EDV30230.1 GGFK01005333 MBW38654.1 CH480815 EDW41945.1 GGFK01004739 MBW38060.1 AE014297 AY051412 AAF55286.3 AAK92836.1 GAMD01001444 JAB00147.1 CP012526 ALC45359.1 GGFL01004766 MBW68944.1 GGFM01001482 MBW22233.1 GGFJ01003900 MBW53041.1 AXCM01018489 GGFM01001401 MBW22152.1 GGFL01004767 MBW68945.1 GGFM01008352 MBW29103.1 DS232244 EDS38307.1 JRES01001582 KNC21762.1 CH933806 EDW15650.1 OUUW01000008 SPP83651.1 SOQ38506.1 LJIG01009436 KRT83083.1 CM000364 EDX12719.1 GANO01002130 JAB57741.1 GFDL01008645 JAV26400.1 GFDL01009959 JAV25086.1 GFDL01008631 JAV26414.1 GFDL01009966 JAV25079.1 GAPW01000803 JAC12795.1 CH940650 EDW66864.1 SPP83652.1 CH477203 EAT48035.1 CH916369 EDV92557.1 AJWK01002452 CCAG010002229 GAKP01004615 JAC54337.1 AJVK01023277 GDHF01023410 JAI28904.1 GBXI01009284 JAD05008.1 GAMC01016458 GAMC01016457 JAB90097.1 GFDF01009199 JAV04885.1 GFDF01009288 JAV04796.1 KPJ06783.1 KQ971312 EEZ98694.2 KQ434984 KZC13133.1 GEBQ01028242 JAT11735.1 JXJN01017337 GECU01037556 JAS70150.1 GECZ01011489 JAS58280.1 CVRI01000047 CRK97916.1 KQ760397 OAD60671.1 GDAI01001882 JAI15721.1 KK107589 QOIP01000001 EZA48635.1 RLU27563.1 KK852463 KDR23392.1 KZ288223 PBC31978.1 GL771642 EFZ09637.1 APGK01053945 KB741239 KB632275 ENN72140.1 ERL91114.1 KQ414710 KOC62989.1 GL446946 EFN87051.1 GEZM01101932 JAV52200.1 GEZM01101933 JAV52199.1
KQ459600 KPI94147.1 KQ461195 KPJ06782.1 AGBW02008253 OWR53915.1 NWSH01003372 PCG66435.1 KA647574 AFP62203.1 ATLV01022206 KE525330 KFB47260.1 GFDG01001231 JAV17568.1 APCN01005901 CH954181 EDV49113.1 AXCN02001770 AAAB01008933 EAA09977.4 CH964272 EDW84202.1 CM000070 EAL28854.1 CM000160 EDW97349.1 CH902623 EDV30230.1 GGFK01005333 MBW38654.1 CH480815 EDW41945.1 GGFK01004739 MBW38060.1 AE014297 AY051412 AAF55286.3 AAK92836.1 GAMD01001444 JAB00147.1 CP012526 ALC45359.1 GGFL01004766 MBW68944.1 GGFM01001482 MBW22233.1 GGFJ01003900 MBW53041.1 AXCM01018489 GGFM01001401 MBW22152.1 GGFL01004767 MBW68945.1 GGFM01008352 MBW29103.1 DS232244 EDS38307.1 JRES01001582 KNC21762.1 CH933806 EDW15650.1 OUUW01000008 SPP83651.1 SOQ38506.1 LJIG01009436 KRT83083.1 CM000364 EDX12719.1 GANO01002130 JAB57741.1 GFDL01008645 JAV26400.1 GFDL01009959 JAV25086.1 GFDL01008631 JAV26414.1 GFDL01009966 JAV25079.1 GAPW01000803 JAC12795.1 CH940650 EDW66864.1 SPP83652.1 CH477203 EAT48035.1 CH916369 EDV92557.1 AJWK01002452 CCAG010002229 GAKP01004615 JAC54337.1 AJVK01023277 GDHF01023410 JAI28904.1 GBXI01009284 JAD05008.1 GAMC01016458 GAMC01016457 JAB90097.1 GFDF01009199 JAV04885.1 GFDF01009288 JAV04796.1 KPJ06783.1 KQ971312 EEZ98694.2 KQ434984 KZC13133.1 GEBQ01028242 JAT11735.1 JXJN01017337 GECU01037556 JAS70150.1 GECZ01011489 JAS58280.1 CVRI01000047 CRK97916.1 KQ760397 OAD60671.1 GDAI01001882 JAI15721.1 KK107589 QOIP01000001 EZA48635.1 RLU27563.1 KK852463 KDR23392.1 KZ288223 PBC31978.1 GL771642 EFZ09637.1 APGK01053945 KB741239 KB632275 ENN72140.1 ERL91114.1 KQ414710 KOC62989.1 GL446946 EFN87051.1 GEZM01101932 JAV52200.1 GEZM01101933 JAV52199.1
Proteomes
UP000005204
UP000037510
UP000053268
UP000053240
UP000007151
UP000218220
+ More
UP000095301 UP000030765 UP000075900 UP000095300 UP000075903 UP000075840 UP000075882 UP000076407 UP000075880 UP000075902 UP000076408 UP000008711 UP000075886 UP000007062 UP000007798 UP000075920 UP000001819 UP000002282 UP000007801 UP000075881 UP000001292 UP000000803 UP000069272 UP000092553 UP000075883 UP000002320 UP000037069 UP000009192 UP000075884 UP000268350 UP000192221 UP000000304 UP000075885 UP000008792 UP000008820 UP000001070 UP000092461 UP000092444 UP000092445 UP000092462 UP000078200 UP000091820 UP000007266 UP000076502 UP000092443 UP000092460 UP000183832 UP000053097 UP000279307 UP000027135 UP000242457 UP000192223 UP000005203 UP000019118 UP000030742 UP000053825 UP000008237
UP000095301 UP000030765 UP000075900 UP000095300 UP000075903 UP000075840 UP000075882 UP000076407 UP000075880 UP000075902 UP000076408 UP000008711 UP000075886 UP000007062 UP000007798 UP000075920 UP000001819 UP000002282 UP000007801 UP000075881 UP000001292 UP000000803 UP000069272 UP000092553 UP000075883 UP000002320 UP000037069 UP000009192 UP000075884 UP000268350 UP000192221 UP000000304 UP000075885 UP000008792 UP000008820 UP000001070 UP000092461 UP000092444 UP000092445 UP000092462 UP000078200 UP000091820 UP000007266 UP000076502 UP000092443 UP000092460 UP000183832 UP000053097 UP000279307 UP000027135 UP000242457 UP000192223 UP000005203 UP000019118 UP000030742 UP000053825 UP000008237
Pfam
PF00083 Sugar_tr
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9JNA9
A0A2H1VCG7
A0A0L7LRI0
A0A194PLE2
A0A194QMN2
A0A212FJK2
+ More
A0A2A4J2Z5 A0A1I8M620 T1PFX4 A0A084WAL6 A0A182RIT6 A0A1I8PLQ6 A0A1L8EFV0 A0A182VJP9 A0A182HYF0 A0A182LPX9 A0A182WUV5 A0A182J953 A0A182UC81 A0A182YF87 B3P0Z4 A0A182QVY2 Q7Q8X0 B4NKZ9 A0A182VT64 Q294V9 B4PR27 B3MTU9 A0A182K4T8 A0A2M4ACZ2 B4HLC5 A0A2M4ABC8 Q961R9 A0A182FTH7 T1DSK2 A0A0M3QX72 A0A2M4CUC2 A0A2M3Z135 A0A2M4BJB2 A0A182MLM8 A0A2M3Z0U5 A0A2M4CUK7 A0A2M3ZKS7 B0X0R5 A0A0L0BNY0 B4KA02 A0A182NDC5 A0A3B0KF16 A0A1W4VFN7 A0A2H1VCJ0 A0A0T6B785 B4QX65 U5EUX6 A0A1Q3FFR9 A0A1Q3FBY2 A0A1Q3FFU4 A0A1Q3FC36 A0A023ETV6 A0A182PJ93 B4LXZ8 A0A3B0JTC4 Q17MW7 H9JNB0 B4JG18 A0A1B0C9K9 A0A1S4H482 A0A1B0GCT6 A0A1A9ZM58 A0A034WJ19 A0A1B0D350 A0A0K8UQD0 A0A0A1X2V9 W8AV18 A0A1L8DER9 A0A1L8DEC5 A0A1A9VAZ8 A0A1A9WHP3 A0A194QMM0 D6WAW4 A0A154PMR9 A0A1B6KJY1 A0A1A9XU90 A0A1B0BNC8 A0A1B6H694 A0A1B6G772 A0A1J1IED5 A0A310SKE6 A0A0K8TNM3 A0A026VYB7 A0A067RHW4 A0A2A3EJV7 A0A1W4XL70 A0A087ZUB5 E9JC16 N6TSC1 A0A0L7QWS4 A0A1W4XLF5 E2BB78 A0A1Y1JWN7 A0A1Y1JSF9
A0A2A4J2Z5 A0A1I8M620 T1PFX4 A0A084WAL6 A0A182RIT6 A0A1I8PLQ6 A0A1L8EFV0 A0A182VJP9 A0A182HYF0 A0A182LPX9 A0A182WUV5 A0A182J953 A0A182UC81 A0A182YF87 B3P0Z4 A0A182QVY2 Q7Q8X0 B4NKZ9 A0A182VT64 Q294V9 B4PR27 B3MTU9 A0A182K4T8 A0A2M4ACZ2 B4HLC5 A0A2M4ABC8 Q961R9 A0A182FTH7 T1DSK2 A0A0M3QX72 A0A2M4CUC2 A0A2M3Z135 A0A2M4BJB2 A0A182MLM8 A0A2M3Z0U5 A0A2M4CUK7 A0A2M3ZKS7 B0X0R5 A0A0L0BNY0 B4KA02 A0A182NDC5 A0A3B0KF16 A0A1W4VFN7 A0A2H1VCJ0 A0A0T6B785 B4QX65 U5EUX6 A0A1Q3FFR9 A0A1Q3FBY2 A0A1Q3FFU4 A0A1Q3FC36 A0A023ETV6 A0A182PJ93 B4LXZ8 A0A3B0JTC4 Q17MW7 H9JNB0 B4JG18 A0A1B0C9K9 A0A1S4H482 A0A1B0GCT6 A0A1A9ZM58 A0A034WJ19 A0A1B0D350 A0A0K8UQD0 A0A0A1X2V9 W8AV18 A0A1L8DER9 A0A1L8DEC5 A0A1A9VAZ8 A0A1A9WHP3 A0A194QMM0 D6WAW4 A0A154PMR9 A0A1B6KJY1 A0A1A9XU90 A0A1B0BNC8 A0A1B6H694 A0A1B6G772 A0A1J1IED5 A0A310SKE6 A0A0K8TNM3 A0A026VYB7 A0A067RHW4 A0A2A3EJV7 A0A1W4XL70 A0A087ZUB5 E9JC16 N6TSC1 A0A0L7QWS4 A0A1W4XLF5 E2BB78 A0A1Y1JWN7 A0A1Y1JSF9
PDB
4LDS
E-value=1.25103e-06,
Score=126
Ontologies
GO
Topology
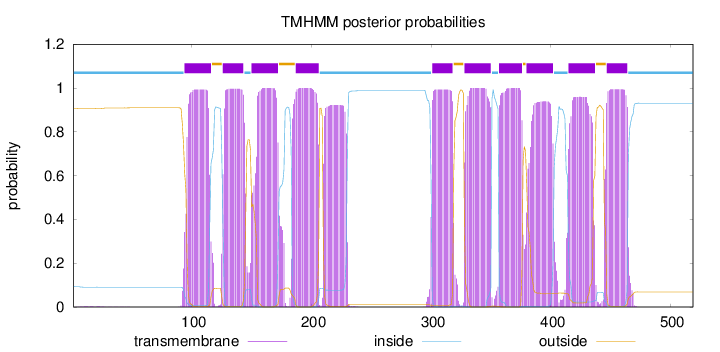
Length:
519
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
227.4628
Exp number, first 60 AAs:
0.06981
Total prob of N-in:
0.09289
inside
1 - 93
TMhelix
94 - 116
outside
117 - 125
TMhelix
126 - 143
inside
144 - 149
TMhelix
150 - 172
outside
173 - 186
TMhelix
187 - 206
inside
207 - 300
TMhelix
301 - 318
outside
319 - 327
TMhelix
328 - 350
inside
351 - 356
TMhelix
357 - 376
outside
377 - 379
TMhelix
380 - 402
inside
403 - 414
TMhelix
415 - 437
outside
438 - 446
TMhelix
447 - 464
inside
465 - 519
Population Genetic Test Statistics
Pi
222.855675
Theta
185.127584
Tajima's D
0.67638
CLR
0.158537
CSRT
0.571421428928554
Interpretation
Uncertain
