Pre Gene Modal
BGIBMGA011014
Annotation
PREDICTED:_histone_deacetylase_complex_subunit_SAP18_[Papilio_machaon]
Full name
Histone deacetylase complex subunit SAP18
+ More
AP-3 complex subunit beta
AP-3 complex subunit beta
Alternative Name
18 kDa Sin3-associated polypeptide
Bicoid-interacting protein 1
dSAP18
Bicoid-interacting protein 1
dSAP18
Location in the cell
Cytoplasmic Reliability : 1.297 Extracellular Reliability : 1.203 Nuclear Reliability : 1.611
Sequence
CDS
ATGGCGGGATTAGAATCAATGGTAGTTGAAGAAATTAAACCGACAGAAAAACCGGTGGACAGAGAAAAGACCTGTCCTCTATTGCTCCGTGTGTTTTGTTCAACTGGCCGACATAATTCTCCTGGAGACTATGTTAGAGGAAACGTACCTCAGAATGAATTACAAATATATACATGGATGGATGCTACACTGCGAGAACTTACAGGACTTGTAAAAGAAGTTAATCCTGAGACGAGACGGAAAGGCACATACTTTGATTTTGCCATTGTGTATCCTGATATGCGTTCCCCTACATATCGTATGAGGGAAATAGGAGTTACATGTTCCGGACAGAGAGGAGGCGATGATAACAAGACACTTTCACAACTGAAATTTCAAATTGGGAACTATCTAGACATCTCAATAACACCCCCAAACAGAATGCCACCACCAATGAGGCGTCCACAACCATATATTAATAATCGTCAGTACTGA
Protein
MAGLESMVVEEIKPTEKPVDREKTCPLLLRVFCSTGRHNSPGDYVRGNVPQNELQIYTWMDATLRELTGLVKEVNPETRRKGTYFDFAIVYPDMRSPTYRMREIGVTCSGQRGGDDNKTLSQLKFQIGNYLDISITPPNRMPPPMRRPQPYINNRQY
Summary
Description
Involved in the tethering of the SIN3 complex to core histone proteins.
Involved in the tethering of the SIN3 complex to core histone proteins. Interacts with bicoid (bcd) to repress transcription of bicoid target genes in the anterior tip of the embryo; a process known as retraction. Interacts with Trl and binds to Polycomb response elements at the bithorax complex. May contribute to the regulation of other homeotic gene expressions.
Involved in the tethering of the SIN3 complex to core histone proteins. Interacts with bicoid (bcd) to repress transcription of bicoid target genes in the anterior tip of the embryo; a process known as retraction. Interacts with Trl and binds to Polycomb response elements at the bithorax complex. May contribute to the regulation of other homeotic gene expressions.
Subunit
Forms a complex with SIN3 and histone deacetylase (By similarity). Interacts with the N-terminal residues of TRL. Interacts with BCD; in vitro and yeast cells.
Similarity
Belongs to the SAP18 family.
Belongs to the adaptor complexes large subunit family.
Belongs to the adaptor complexes large subunit family.
Keywords
Complete proteome
Cytoplasm
Developmental protein
Nucleus
Reference proteome
Repressor
Transcription
Transcription regulation
Feature
chain Histone deacetylase complex subunit SAP18
Uniprot
H9JNB1
A0A2H1VCD3
A0A212FJJ4
A0A2A4J8H3
S4PA87
A0A0L7LS92
+ More
A0A1E1WB42 A0A194QP84 I4DLA8 A0A2J7RBZ4 A0A084VRE8 A0A182MMQ9 A0A182SMH3 A0A182NFN1 A0A182RNP0 A0A182QS34 A0A0K8TSH3 A0A182Y4D0 A0A182K2Q1 A0A182IZ79 A0A182LFR3 A0A182TEW6 A0A182HUA2 Q7PR28 A0A210Q1W2 E9IJQ7 A0A023EGW8 A0A2M3ZD68 A0A182F7F7 A0A2M4C2H2 A0A158ND33 A0A151XGE8 E2B7E5 A0A026VXI5 A0A195DUK5 F4X0G6 A0A195F7M0 A0A2M4AYH4 W5JHI1 A0A154PLG4 A0A2A3E3B0 A0A1Q3F8N6 A0A0L7QP96 B0XBD8 A0A1B0GCT3 A0A1A9XU91 A0A1A9VB00 A0A1B0BND1 A0A1A9ZM65 A0A1A9WHP5 A0A151ING1 B0XKQ7 A0A1J1HX58 A0A0L0BNU0 A0A182PAB1 A0A1I8N9K0 U5EXK6 A0A1B0C9K6 A0A1I8PWK3 A0A1L8DGB9 T1EA26 W8AXT2 A0A0K8VWW1 A0A034V189 B4QX67 E2A025 T1IHD1 B4PR29 B3P0Z2 B4HLC7 A0A126GUU7 Q9VEX9 A0A310SA59 A0A0N0U7P8 A0A232F990 K7JAE3 A0A182W084 B4GMJ9 A0A087TH09 V4A2F2 B4KA04 B4JG20 B4LY00 A0A0B7BRP6 A0A182H595 A0A2C9JZ69 A0A1W4VU25 A0A3B0JN61 B3LZX9 A0A0M4F2M6 T1IKX6 Q294V7 K1QH12 T1JKW2 B4NL01 A0A088AMR2 J9Q660 A0A1Q3F8G7 A0A0L8HF97 Q09JJ5
A0A1E1WB42 A0A194QP84 I4DLA8 A0A2J7RBZ4 A0A084VRE8 A0A182MMQ9 A0A182SMH3 A0A182NFN1 A0A182RNP0 A0A182QS34 A0A0K8TSH3 A0A182Y4D0 A0A182K2Q1 A0A182IZ79 A0A182LFR3 A0A182TEW6 A0A182HUA2 Q7PR28 A0A210Q1W2 E9IJQ7 A0A023EGW8 A0A2M3ZD68 A0A182F7F7 A0A2M4C2H2 A0A158ND33 A0A151XGE8 E2B7E5 A0A026VXI5 A0A195DUK5 F4X0G6 A0A195F7M0 A0A2M4AYH4 W5JHI1 A0A154PLG4 A0A2A3E3B0 A0A1Q3F8N6 A0A0L7QP96 B0XBD8 A0A1B0GCT3 A0A1A9XU91 A0A1A9VB00 A0A1B0BND1 A0A1A9ZM65 A0A1A9WHP5 A0A151ING1 B0XKQ7 A0A1J1HX58 A0A0L0BNU0 A0A182PAB1 A0A1I8N9K0 U5EXK6 A0A1B0C9K6 A0A1I8PWK3 A0A1L8DGB9 T1EA26 W8AXT2 A0A0K8VWW1 A0A034V189 B4QX67 E2A025 T1IHD1 B4PR29 B3P0Z2 B4HLC7 A0A126GUU7 Q9VEX9 A0A310SA59 A0A0N0U7P8 A0A232F990 K7JAE3 A0A182W084 B4GMJ9 A0A087TH09 V4A2F2 B4KA04 B4JG20 B4LY00 A0A0B7BRP6 A0A182H595 A0A2C9JZ69 A0A1W4VU25 A0A3B0JN61 B3LZX9 A0A0M4F2M6 T1IKX6 Q294V7 K1QH12 T1JKW2 B4NL01 A0A088AMR2 J9Q660 A0A1Q3F8G7 A0A0L8HF97 Q09JJ5
Pubmed
19121390
22118469
23622113
26227816
26354079
22651552
+ More
24438588 26369729 25244985 20966253 12364791 14747013 17210077 28812685 21282665 24945155 21347285 20798317 24508170 30249741 21719571 20920257 23761445 26108605 25315136 24495485 25348373 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 11455422 12537569 11256608 28648823 20075255 23254933 26483478 15562597 15632085 22992520 22833317 18070664
24438588 26369729 25244985 20966253 12364791 14747013 17210077 28812685 21282665 24945155 21347285 20798317 24508170 30249741 21719571 20920257 23761445 26108605 25315136 24495485 25348373 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 11455422 12537569 11256608 28648823 20075255 23254933 26483478 15562597 15632085 22992520 22833317 18070664
EMBL
BABH01019612
ODYU01001804
SOQ38508.1
AGBW02008253
OWR53913.1
NWSH01002561
+ More
PCG67978.1 GAIX01003359 JAA89201.1 JTDY01000250 KOB78066.1 GDQN01006842 JAT84212.1 KQ461195 KPJ06785.1 AK402076 KQ459600 BAM18698.1 KPI94143.1 NEVH01005889 PNF38349.1 ATLV01015610 KE525023 KFB40542.1 AXCM01000851 AXCN02000239 GDAI01000291 JAI17312.1 APCN01000613 AAAB01008859 EAA07931.3 NEDP02005224 OWF42743.1 GL763883 EFZ19150.1 GAPW01005512 JAC08086.1 GGFM01005733 MBW26484.1 GGFJ01010352 MBW59493.1 ADTU01012291 KQ982173 KYQ59381.1 GL446169 EFN88381.1 KK107609 QOIP01000014 EZA48518.1 RLU15103.1 KQ980341 KYN16437.1 GL888498 EGI59958.1 KQ981744 KYN36438.1 GGFK01012510 MBW45831.1 ADMH02001371 ETN62768.1 KQ434938 KZC12070.1 KZ288433 PBC25782.1 GFDL01011105 JAV23940.1 KQ414843 KOC60321.1 DS232629 EDS44229.1 CCAG010002226 JXJN01017337 KQ976933 KYN07002.1 DS233966 EDS32453.1 CVRI01000020 CRK91110.1 JRES01001582 KNC21755.1 GANO01000907 JAB58964.1 AJWK01002450 GFDF01008576 JAV05508.1 GAMD01001383 JAB00208.1 GAMC01012760 JAB93795.1 GDHF01008946 JAI43368.1 GAKP01022698 JAC36254.1 CM000364 EDX12721.1 GL435543 EFN73208.1 JH429868 CM000160 EDW97351.1 CH954181 EDV49111.1 CH480815 EDW41947.1 AE014297 KX671631 ALI30577.1 AOG29656.1 AF297546 AY070913 BT025229 AJ278500 KQ767242 OAD53481.1 KQ435689 KOX81302.1 NNAY01000604 OXU27424.1 AAZX01004373 CH479185 EDW38073.1 KK115199 KFM64398.1 KB202619 ESO89120.1 CH933806 EDW15652.1 CH916369 EDV92559.1 CH940650 EDW66866.1 HACG01048692 CEK95557.1 JXUM01110979 JXUM01110980 JXUM01110981 JXUM01110982 JXUM01110983 JXUM01110984 KQ565461 KXJ70939.1 OUUW01000008 SPP83654.1 CH902617 EDV44169.1 CP012526 ALC45357.1 JH430667 CM000070 EAL28856.1 JH818298 EKC36057.1 JH431633 CH964272 EDW84204.1 JN184771 AFK73705.1 GFDL01011209 JAV23836.1 KQ418297 KOF87911.1 DQ886851 ABI52768.1
PCG67978.1 GAIX01003359 JAA89201.1 JTDY01000250 KOB78066.1 GDQN01006842 JAT84212.1 KQ461195 KPJ06785.1 AK402076 KQ459600 BAM18698.1 KPI94143.1 NEVH01005889 PNF38349.1 ATLV01015610 KE525023 KFB40542.1 AXCM01000851 AXCN02000239 GDAI01000291 JAI17312.1 APCN01000613 AAAB01008859 EAA07931.3 NEDP02005224 OWF42743.1 GL763883 EFZ19150.1 GAPW01005512 JAC08086.1 GGFM01005733 MBW26484.1 GGFJ01010352 MBW59493.1 ADTU01012291 KQ982173 KYQ59381.1 GL446169 EFN88381.1 KK107609 QOIP01000014 EZA48518.1 RLU15103.1 KQ980341 KYN16437.1 GL888498 EGI59958.1 KQ981744 KYN36438.1 GGFK01012510 MBW45831.1 ADMH02001371 ETN62768.1 KQ434938 KZC12070.1 KZ288433 PBC25782.1 GFDL01011105 JAV23940.1 KQ414843 KOC60321.1 DS232629 EDS44229.1 CCAG010002226 JXJN01017337 KQ976933 KYN07002.1 DS233966 EDS32453.1 CVRI01000020 CRK91110.1 JRES01001582 KNC21755.1 GANO01000907 JAB58964.1 AJWK01002450 GFDF01008576 JAV05508.1 GAMD01001383 JAB00208.1 GAMC01012760 JAB93795.1 GDHF01008946 JAI43368.1 GAKP01022698 JAC36254.1 CM000364 EDX12721.1 GL435543 EFN73208.1 JH429868 CM000160 EDW97351.1 CH954181 EDV49111.1 CH480815 EDW41947.1 AE014297 KX671631 ALI30577.1 AOG29656.1 AF297546 AY070913 BT025229 AJ278500 KQ767242 OAD53481.1 KQ435689 KOX81302.1 NNAY01000604 OXU27424.1 AAZX01004373 CH479185 EDW38073.1 KK115199 KFM64398.1 KB202619 ESO89120.1 CH933806 EDW15652.1 CH916369 EDV92559.1 CH940650 EDW66866.1 HACG01048692 CEK95557.1 JXUM01110979 JXUM01110980 JXUM01110981 JXUM01110982 JXUM01110983 JXUM01110984 KQ565461 KXJ70939.1 OUUW01000008 SPP83654.1 CH902617 EDV44169.1 CP012526 ALC45357.1 JH430667 CM000070 EAL28856.1 JH818298 EKC36057.1 JH431633 CH964272 EDW84204.1 JN184771 AFK73705.1 GFDL01011209 JAV23836.1 KQ418297 KOF87911.1 DQ886851 ABI52768.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000037510
UP000053240
UP000053268
+ More
UP000235965 UP000030765 UP000075883 UP000075901 UP000075884 UP000075900 UP000075886 UP000076408 UP000075881 UP000075880 UP000075882 UP000075902 UP000075840 UP000007062 UP000242188 UP000069272 UP000005205 UP000075809 UP000008237 UP000053097 UP000279307 UP000078492 UP000007755 UP000078541 UP000000673 UP000076502 UP000242457 UP000053825 UP000002320 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000078542 UP000183832 UP000037069 UP000075885 UP000095301 UP000092461 UP000095300 UP000000304 UP000000311 UP000002282 UP000008711 UP000001292 UP000000803 UP000053105 UP000215335 UP000002358 UP000075920 UP000008744 UP000054359 UP000030746 UP000009192 UP000001070 UP000008792 UP000069940 UP000249989 UP000076420 UP000192221 UP000268350 UP000007801 UP000092553 UP000001819 UP000005408 UP000007798 UP000005203 UP000053454
UP000235965 UP000030765 UP000075883 UP000075901 UP000075884 UP000075900 UP000075886 UP000076408 UP000075881 UP000075880 UP000075882 UP000075902 UP000075840 UP000007062 UP000242188 UP000069272 UP000005205 UP000075809 UP000008237 UP000053097 UP000279307 UP000078492 UP000007755 UP000078541 UP000000673 UP000076502 UP000242457 UP000053825 UP000002320 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000091820 UP000078542 UP000183832 UP000037069 UP000075885 UP000095301 UP000092461 UP000095300 UP000000304 UP000000311 UP000002282 UP000008711 UP000001292 UP000000803 UP000053105 UP000215335 UP000002358 UP000075920 UP000008744 UP000054359 UP000030746 UP000009192 UP000001070 UP000008792 UP000069940 UP000249989 UP000076420 UP000192221 UP000268350 UP000007801 UP000092553 UP000001819 UP000005408 UP000007798 UP000005203 UP000053454
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
CDD
ProteinModelPortal
H9JNB1
A0A2H1VCD3
A0A212FJJ4
A0A2A4J8H3
S4PA87
A0A0L7LS92
+ More
A0A1E1WB42 A0A194QP84 I4DLA8 A0A2J7RBZ4 A0A084VRE8 A0A182MMQ9 A0A182SMH3 A0A182NFN1 A0A182RNP0 A0A182QS34 A0A0K8TSH3 A0A182Y4D0 A0A182K2Q1 A0A182IZ79 A0A182LFR3 A0A182TEW6 A0A182HUA2 Q7PR28 A0A210Q1W2 E9IJQ7 A0A023EGW8 A0A2M3ZD68 A0A182F7F7 A0A2M4C2H2 A0A158ND33 A0A151XGE8 E2B7E5 A0A026VXI5 A0A195DUK5 F4X0G6 A0A195F7M0 A0A2M4AYH4 W5JHI1 A0A154PLG4 A0A2A3E3B0 A0A1Q3F8N6 A0A0L7QP96 B0XBD8 A0A1B0GCT3 A0A1A9XU91 A0A1A9VB00 A0A1B0BND1 A0A1A9ZM65 A0A1A9WHP5 A0A151ING1 B0XKQ7 A0A1J1HX58 A0A0L0BNU0 A0A182PAB1 A0A1I8N9K0 U5EXK6 A0A1B0C9K6 A0A1I8PWK3 A0A1L8DGB9 T1EA26 W8AXT2 A0A0K8VWW1 A0A034V189 B4QX67 E2A025 T1IHD1 B4PR29 B3P0Z2 B4HLC7 A0A126GUU7 Q9VEX9 A0A310SA59 A0A0N0U7P8 A0A232F990 K7JAE3 A0A182W084 B4GMJ9 A0A087TH09 V4A2F2 B4KA04 B4JG20 B4LY00 A0A0B7BRP6 A0A182H595 A0A2C9JZ69 A0A1W4VU25 A0A3B0JN61 B3LZX9 A0A0M4F2M6 T1IKX6 Q294V7 K1QH12 T1JKW2 B4NL01 A0A088AMR2 J9Q660 A0A1Q3F8G7 A0A0L8HF97 Q09JJ5
A0A1E1WB42 A0A194QP84 I4DLA8 A0A2J7RBZ4 A0A084VRE8 A0A182MMQ9 A0A182SMH3 A0A182NFN1 A0A182RNP0 A0A182QS34 A0A0K8TSH3 A0A182Y4D0 A0A182K2Q1 A0A182IZ79 A0A182LFR3 A0A182TEW6 A0A182HUA2 Q7PR28 A0A210Q1W2 E9IJQ7 A0A023EGW8 A0A2M3ZD68 A0A182F7F7 A0A2M4C2H2 A0A158ND33 A0A151XGE8 E2B7E5 A0A026VXI5 A0A195DUK5 F4X0G6 A0A195F7M0 A0A2M4AYH4 W5JHI1 A0A154PLG4 A0A2A3E3B0 A0A1Q3F8N6 A0A0L7QP96 B0XBD8 A0A1B0GCT3 A0A1A9XU91 A0A1A9VB00 A0A1B0BND1 A0A1A9ZM65 A0A1A9WHP5 A0A151ING1 B0XKQ7 A0A1J1HX58 A0A0L0BNU0 A0A182PAB1 A0A1I8N9K0 U5EXK6 A0A1B0C9K6 A0A1I8PWK3 A0A1L8DGB9 T1EA26 W8AXT2 A0A0K8VWW1 A0A034V189 B4QX67 E2A025 T1IHD1 B4PR29 B3P0Z2 B4HLC7 A0A126GUU7 Q9VEX9 A0A310SA59 A0A0N0U7P8 A0A232F990 K7JAE3 A0A182W084 B4GMJ9 A0A087TH09 V4A2F2 B4KA04 B4JG20 B4LY00 A0A0B7BRP6 A0A182H595 A0A2C9JZ69 A0A1W4VU25 A0A3B0JN61 B3LZX9 A0A0M4F2M6 T1IKX6 Q294V7 K1QH12 T1JKW2 B4NL01 A0A088AMR2 J9Q660 A0A1Q3F8G7 A0A0L8HF97 Q09JJ5
PDB
4A90
E-value=7.65276e-43,
Score=432
Ontologies
PATHWAY
GO
Topology
Subcellular location
Nucleus
Shuttles between the nucleus and the cytoplasm. With evidence from 2 publications.
Cytoplasm Shuttles between the nucleus and the cytoplasm. With evidence from 2 publications.
Cytoplasm Shuttles between the nucleus and the cytoplasm. With evidence from 2 publications.
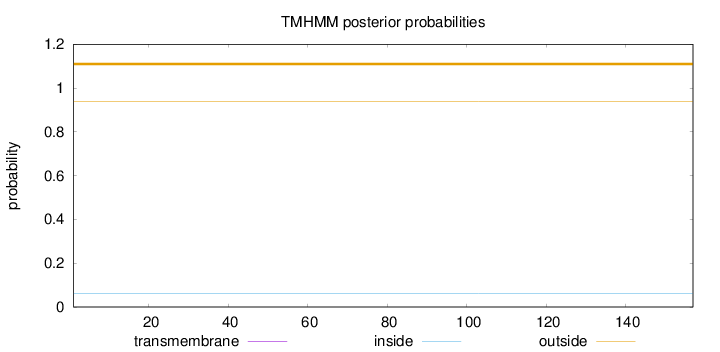
Length:
157
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0
Exp number, first 60 AAs:
0
Total prob of N-in:
0.06210
outside
1 - 157
Population Genetic Test Statistics
Pi
411.321213
Theta
228.284882
Tajima's D
2.524996
CLR
0.493795
CSRT
0.947152642367882
Interpretation
Uncertain
