Gene
KWMTBOMO13705
Pre Gene Modal
BGIBMGA011093
Annotation
PREDICTED:_organic_cation_transporter_protein-like_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.982
Sequence
CDS
ATGACGTTCCTACTCTCCCTGTTCTCATTGCCATGCACGTTTCATATTTACCTGCCTACTTTTACTGCAAAAGCAACTAAATTCTGGTGCAGTAGGCCCGACAATTTGTCAAATTTGTCAATAACCGAATGGATCAACTACAGCCAGCCGATCAACGCTTGCAGTGTACGAAAACTGCCTCCAGGAATAACAGCTGAAGCGATACTCAATCACACTGCTCCAATTTTGGCAAATTTATTCGAAAAATGTACTGAATGGGAGTACGATAGATCAGAAGTAGGGAACACAATAATAACCGAATGGAACTTGGTGTGTGATAACGCAAGTCTTACGAGTTTGGCTGAAGTCGTCTTTTTGATAGGAATTGGCGTAGGTGGAGTTATCGGAGGATGGATTTCAGACAGATTCGGACGTAAACGTATTTTGATGGGCATGATAACAGCGCAAAGCGTTTTGGCCATTCTGTCACTACTGGTCAGGTCATACGTCCAATATGTGGTCGTAAGATTGATTATGGGGTTCGTCTCCGTGTCAGTGGTGTATGCAGCTTTTGTTCTTTCTGTAGAACTCGTCGGTGGCAAATGGGTCACCATTGCTGGCGTGTGCAACTTCTTTCCCTTGCCACTGGCGTACCTCATTGTCTCTCTGTTGTCTTTAGTCCTACCAAATTGGAGAGATCTACAACTGGCATTGTCGATACCCGGGTGCCTATTGATTGCGCTATGGTTTGTTTTGCCGGAGTCACCAAGATGGCTACTGAGCATGGGAAAAACTCAAGAAGCGAAACTGATGTTGCAAAAAGCAGCTATGTTCAACAAACGAGAGCTACCCGCGGATATCGACAAGCTGTTGTTATTACAAAAATCGGAGACACGAGAAGAAGGTCCCAGCGTTACTATGCTCTTTAAGGGACCTTTACGCAAACGGACGATTTGTTTATTTGTAGCTTGGTTTTGTATGACCATTGTGTACTATGGACTATTATTGAACATTGGGAATTTCAACATAGGAAATCTCCACATGACATCAATAATCCTGGCAATAGTCGAGATCCCTGCCATTGCCGTGAGTGTACCCATTCTTTTAAAAGGAGGGAGACGAATTCCAATTGTCAGCAGTATGATAATATGTGGTCTGGCCTGTGTGGCCAGCGAATTATTCTCGATAGCGTTTGAACAAGAATGGGTGATGATATTGTGTTTGATGGTTGGAAAATTTGCTATTGGATCAACGAACATGATGTTACCGATATACACTGTGGAACTATATCCAACCGTAGTGAGAAATTTGGGCGTTGGAGCGAGCCAAATGTCAGCTGGTTTGGCATTGATTTTCATTCCGTATCTGTGGAAATTGGCAGCACTGAATGAACATTTACCGATGGTCACAATATCAGCTTTAGGGGCAATCGGCGGTGGCATTGTGCTAATCCTTCCGGAAACGGGACAGTTGAAAGAGCCACCAGCGACTGTTGTTGGAAAATTACCGCAATATGACAGCAACTCTCCTTGCAACGGAACGTTTACGGTCGCCGACGAGAGAGGGCATTAG
Protein
MTFLLSLFSLPCTFHIYLPTFTAKATKFWCSRPDNLSNLSITEWINYSQPINACSVRKLPPGITAEAILNHTAPILANLFEKCTEWEYDRSEVGNTIITEWNLVCDNASLTSLAEVVFLIGIGVGGVIGGWISDRFGRKRILMGMITAQSVLAILSLLVRSYVQYVVVRLIMGFVSVSVVYAAFVLSVELVGGKWVTIAGVCNFFPLPLAYLIVSLLSLVLPNWRDLQLALSIPGCLLIALWFVLPESPRWLLSMGKTQEAKLMLQKAAMFNKRELPADIDKLLLLQKSETREEGPSVTMLFKGPLRKRTICLFVAWFCMTIVYYGLLLNIGNFNIGNLHMTSIILAIVEIPAIAVSVPILLKGGRRIPIVSSMIICGLACVASELFSIAFEQEWVMILCLMVGKFAIGSTNMMLPIYTVELYPTVVRNLGVGASQMSAGLALIFIPYLWKLAALNEHLPMVTISALGAIGGGIVLILPETGQLKEPPATVVGKLPQYDSNSPCNGTFTVADERGH
Summary
Uniprot
H9JNI9
A0A2H1VCD7
A0A2A4J886
A0A194PL27
A0A3S2LVG4
A0A194QMN8
+ More
A0A0L7LRP1 D6W9G4 A0A2J7RC21 A0A1S3DM74 A0A1Y1MY32 A0A1Y1MXZ1 A0A2R7WEP4 A0A023F2X8 V5GHW6 A0A1B6MRI4 T1HMH5 N6TDH0 E0VD76 A0A0T6BHL7 A0A1B6J9F7 A0A2S2R8L8 A0A067RG97 J9JKH9 A0A2H8TNI1 B4NL04 A0A3Q0IMQ1 A0A2S2PEK9 A0A336M2N9 A0A146LZE2 A0A146LK24 A0A146LBD8 B0X4Q2 A0A182QQK7 A0A2M4BK64 A0A182NFM7 A0A182K8I9 A0A182Y4D4 A0A182W088 A0A2M4BIP6 A0A1L8DDY6 A0A182PAB5 F5HKA0 A0A2M4BHC1 A0A182MJ90 B4GMK2 A0A182RNN6 I5APN8 A0A1Y9INC2 W5JEI9 B5DW31 A0A1C7CZB1 A0A1S4HC19 A0A3F2Z0C8 A0A1Y9GLM9 A0A1I8JVP7 A0A0P6J4H9 Q17CS3 A0A0M4ECH5 Q8MQI2 B4QX70 B4HLD0 A0A0R1E318 A2RVG5 A0A0Q9X0P4 B4JG23 B3P0Y9 A0A3B0KIU3 B4KAE2 B4PR32 A0A0Q9XBC6 A0A1W4VVS4 A0A3B0JN53 Q9VEY2 A0A182F7G1 A0A1J1IAV1 A0A1W4VU24 A0A0P9ATM0 A0A182JKT0 A0A1J1ICR0 B3LZY2 B4LY03 A0A182QAD4 A0A0Q9WRP9 A0A1I8PD63 A0A1I8MGW3 W8BB78 W8ARB1 A0A1I8MGW6 T1PHN4 A0A1B0FML0 A0A1B0C9K4 A0A182V4P1 A0A182II23 Q7QDB1 A0A182LFQ7 A0A0A1WVX1 A0A1A9VAY2 A0A1A9W3U2 A0A1Q3G1I7
A0A0L7LRP1 D6W9G4 A0A2J7RC21 A0A1S3DM74 A0A1Y1MY32 A0A1Y1MXZ1 A0A2R7WEP4 A0A023F2X8 V5GHW6 A0A1B6MRI4 T1HMH5 N6TDH0 E0VD76 A0A0T6BHL7 A0A1B6J9F7 A0A2S2R8L8 A0A067RG97 J9JKH9 A0A2H8TNI1 B4NL04 A0A3Q0IMQ1 A0A2S2PEK9 A0A336M2N9 A0A146LZE2 A0A146LK24 A0A146LBD8 B0X4Q2 A0A182QQK7 A0A2M4BK64 A0A182NFM7 A0A182K8I9 A0A182Y4D4 A0A182W088 A0A2M4BIP6 A0A1L8DDY6 A0A182PAB5 F5HKA0 A0A2M4BHC1 A0A182MJ90 B4GMK2 A0A182RNN6 I5APN8 A0A1Y9INC2 W5JEI9 B5DW31 A0A1C7CZB1 A0A1S4HC19 A0A3F2Z0C8 A0A1Y9GLM9 A0A1I8JVP7 A0A0P6J4H9 Q17CS3 A0A0M4ECH5 Q8MQI2 B4QX70 B4HLD0 A0A0R1E318 A2RVG5 A0A0Q9X0P4 B4JG23 B3P0Y9 A0A3B0KIU3 B4KAE2 B4PR32 A0A0Q9XBC6 A0A1W4VVS4 A0A3B0JN53 Q9VEY2 A0A182F7G1 A0A1J1IAV1 A0A1W4VU24 A0A0P9ATM0 A0A182JKT0 A0A1J1ICR0 B3LZY2 B4LY03 A0A182QAD4 A0A0Q9WRP9 A0A1I8PD63 A0A1I8MGW3 W8BB78 W8ARB1 A0A1I8MGW6 T1PHN4 A0A1B0FML0 A0A1B0C9K4 A0A182V4P1 A0A182II23 Q7QDB1 A0A182LFQ7 A0A0A1WVX1 A0A1A9VAY2 A0A1A9W3U2 A0A1Q3G1I7
Pubmed
19121390
26354079
26227816
18362917
19820115
28004739
+ More
25474469 23537049 20566863 24845553 17994087 26823975 25244985 12364791 15632085 23185243 20920257 23761445 20966253 26999592 17510324 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 25315136 24495485 14747013 17210077 25830018
25474469 23537049 20566863 24845553 17994087 26823975 25244985 12364791 15632085 23185243 20920257 23761445 20966253 26999592 17510324 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 18057021 25315136 24495485 14747013 17210077 25830018
EMBL
BABH01019609
ODYU01001804
SOQ38510.1
NWSH01002561
PCG67976.1
KQ459600
+ More
KPI94141.1 RSAL01000184 RVE44862.1 KQ461195 KPJ06787.1 JTDY01000250 KOB78064.1 KQ971312 EEZ98159.1 NEVH01005889 PNF38358.1 GEZM01017612 GEZM01017611 JAV90549.1 GEZM01017615 GEZM01017606 JAV90552.1 KK854719 PTY18137.1 GBBI01003144 JAC15568.1 GALX01004832 JAB63634.1 GEBQ01001468 JAT38509.1 ACPB03001266 APGK01042252 APGK01042253 APGK01042254 APGK01042255 KB741002 KB632309 ENN75788.1 ERL92047.1 DS235072 EEB11332.1 LJIG01000136 KRT86804.1 GECU01011973 GECU01010821 JAS95733.1 JAS96885.1 GGMS01017163 MBY86366.1 KK852482 KDR22901.1 ABLF02024820 ABLF02024821 ABLF02024822 ABLF02024823 ABLF02024824 GFXV01003387 MBW15192.1 CH964272 EDW84207.1 GGMR01014727 MBY27346.1 UFQS01003907 UFQT01000449 UFQT01003907 SSX16028.1 SSX24482.1 GDHC01011738 GDHC01005671 JAQ06891.1 JAQ12958.1 GDHC01010248 JAQ08381.1 GDHC01013011 JAQ05618.1 DS232350 EDS40469.1 AXCN02000238 GGFJ01004328 MBW53469.1 GGFJ01003733 MBW52874.1 GFDF01009413 JAV04671.1 AAAB01008859 EGK96652.1 GGFJ01003210 MBW52351.1 AXCM01000851 CH479185 EDW38076.1 CM000070 EIM52923.1 KRT01124.1 ADMH02001371 ETN62772.1 EDY68499.1 APCN01000614 GDUN01000920 JAN94999.1 CH477305 EAT44122.1 CP012526 ALC45354.1 AY129463 AAM76205.1 CM000364 EDX12724.1 CH480815 EDW41950.1 CM000160 KRK03669.1 KRK03670.1 KRK03671.1 BT029956 AE014297 ABM92830.1 ABW08686.1 CH933806 KRG01656.1 KRG01657.1 KRG01658.1 KRG01659.1 CH916369 EDV92562.1 CH954181 EDV49108.1 OUUW01000008 SPP83658.1 EDW15655.2 EDW97354.2 KRG01655.1 SPP83657.1 AAF55281.1 AFH06450.1 AFH06451.1 AFH06452.1 CVRI01000047 CRK97339.1 CH902617 KPU80647.1 KPU80648.1 KPU80649.1 KPU80650.1 KPU80651.1 KPU80652.1 CRK97340.1 EDV44172.1 CH940650 EDW66869.1 KRF82949.1 GAMC01019456 JAB87099.1 GAMC01019457 GAMC01019455 JAB87098.1 KA648332 AFP62961.1 CCAG010015076 AJWK01002444 AJWK01002445 AJWK01002446 AJWK01002447 AJWK01002448 APCN01000613 EAA07911.5 GBXI01011714 JAD02578.1 GFDL01001399 JAV33646.1
KPI94141.1 RSAL01000184 RVE44862.1 KQ461195 KPJ06787.1 JTDY01000250 KOB78064.1 KQ971312 EEZ98159.1 NEVH01005889 PNF38358.1 GEZM01017612 GEZM01017611 JAV90549.1 GEZM01017615 GEZM01017606 JAV90552.1 KK854719 PTY18137.1 GBBI01003144 JAC15568.1 GALX01004832 JAB63634.1 GEBQ01001468 JAT38509.1 ACPB03001266 APGK01042252 APGK01042253 APGK01042254 APGK01042255 KB741002 KB632309 ENN75788.1 ERL92047.1 DS235072 EEB11332.1 LJIG01000136 KRT86804.1 GECU01011973 GECU01010821 JAS95733.1 JAS96885.1 GGMS01017163 MBY86366.1 KK852482 KDR22901.1 ABLF02024820 ABLF02024821 ABLF02024822 ABLF02024823 ABLF02024824 GFXV01003387 MBW15192.1 CH964272 EDW84207.1 GGMR01014727 MBY27346.1 UFQS01003907 UFQT01000449 UFQT01003907 SSX16028.1 SSX24482.1 GDHC01011738 GDHC01005671 JAQ06891.1 JAQ12958.1 GDHC01010248 JAQ08381.1 GDHC01013011 JAQ05618.1 DS232350 EDS40469.1 AXCN02000238 GGFJ01004328 MBW53469.1 GGFJ01003733 MBW52874.1 GFDF01009413 JAV04671.1 AAAB01008859 EGK96652.1 GGFJ01003210 MBW52351.1 AXCM01000851 CH479185 EDW38076.1 CM000070 EIM52923.1 KRT01124.1 ADMH02001371 ETN62772.1 EDY68499.1 APCN01000614 GDUN01000920 JAN94999.1 CH477305 EAT44122.1 CP012526 ALC45354.1 AY129463 AAM76205.1 CM000364 EDX12724.1 CH480815 EDW41950.1 CM000160 KRK03669.1 KRK03670.1 KRK03671.1 BT029956 AE014297 ABM92830.1 ABW08686.1 CH933806 KRG01656.1 KRG01657.1 KRG01658.1 KRG01659.1 CH916369 EDV92562.1 CH954181 EDV49108.1 OUUW01000008 SPP83658.1 EDW15655.2 EDW97354.2 KRG01655.1 SPP83657.1 AAF55281.1 AFH06450.1 AFH06451.1 AFH06452.1 CVRI01000047 CRK97339.1 CH902617 KPU80647.1 KPU80648.1 KPU80649.1 KPU80650.1 KPU80651.1 KPU80652.1 CRK97340.1 EDV44172.1 CH940650 EDW66869.1 KRF82949.1 GAMC01019456 JAB87099.1 GAMC01019457 GAMC01019455 JAB87098.1 KA648332 AFP62961.1 CCAG010015076 AJWK01002444 AJWK01002445 AJWK01002446 AJWK01002447 AJWK01002448 APCN01000613 EAA07911.5 GBXI01011714 JAD02578.1 GFDL01001399 JAV33646.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000283053
UP000053240
UP000037510
+ More
UP000007266 UP000235965 UP000079169 UP000015103 UP000019118 UP000030742 UP000009046 UP000027135 UP000007819 UP000007798 UP000002320 UP000075886 UP000075884 UP000075881 UP000076408 UP000075920 UP000075885 UP000007062 UP000075883 UP000008744 UP000075900 UP000001819 UP000075902 UP000000673 UP000075882 UP000075903 UP000075840 UP000076407 UP000008820 UP000092553 UP000000304 UP000001292 UP000002282 UP000000803 UP000009192 UP000001070 UP000008711 UP000268350 UP000192221 UP000069272 UP000183832 UP000007801 UP000075880 UP000008792 UP000095300 UP000095301 UP000092444 UP000092461 UP000078200 UP000091820
UP000007266 UP000235965 UP000079169 UP000015103 UP000019118 UP000030742 UP000009046 UP000027135 UP000007819 UP000007798 UP000002320 UP000075886 UP000075884 UP000075881 UP000076408 UP000075920 UP000075885 UP000007062 UP000075883 UP000008744 UP000075900 UP000001819 UP000075902 UP000000673 UP000075882 UP000075903 UP000075840 UP000076407 UP000008820 UP000092553 UP000000304 UP000001292 UP000002282 UP000000803 UP000009192 UP000001070 UP000008711 UP000268350 UP000192221 UP000069272 UP000183832 UP000007801 UP000075880 UP000008792 UP000095300 UP000095301 UP000092444 UP000092461 UP000078200 UP000091820
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9JNI9
A0A2H1VCD7
A0A2A4J886
A0A194PL27
A0A3S2LVG4
A0A194QMN8
+ More
A0A0L7LRP1 D6W9G4 A0A2J7RC21 A0A1S3DM74 A0A1Y1MY32 A0A1Y1MXZ1 A0A2R7WEP4 A0A023F2X8 V5GHW6 A0A1B6MRI4 T1HMH5 N6TDH0 E0VD76 A0A0T6BHL7 A0A1B6J9F7 A0A2S2R8L8 A0A067RG97 J9JKH9 A0A2H8TNI1 B4NL04 A0A3Q0IMQ1 A0A2S2PEK9 A0A336M2N9 A0A146LZE2 A0A146LK24 A0A146LBD8 B0X4Q2 A0A182QQK7 A0A2M4BK64 A0A182NFM7 A0A182K8I9 A0A182Y4D4 A0A182W088 A0A2M4BIP6 A0A1L8DDY6 A0A182PAB5 F5HKA0 A0A2M4BHC1 A0A182MJ90 B4GMK2 A0A182RNN6 I5APN8 A0A1Y9INC2 W5JEI9 B5DW31 A0A1C7CZB1 A0A1S4HC19 A0A3F2Z0C8 A0A1Y9GLM9 A0A1I8JVP7 A0A0P6J4H9 Q17CS3 A0A0M4ECH5 Q8MQI2 B4QX70 B4HLD0 A0A0R1E318 A2RVG5 A0A0Q9X0P4 B4JG23 B3P0Y9 A0A3B0KIU3 B4KAE2 B4PR32 A0A0Q9XBC6 A0A1W4VVS4 A0A3B0JN53 Q9VEY2 A0A182F7G1 A0A1J1IAV1 A0A1W4VU24 A0A0P9ATM0 A0A182JKT0 A0A1J1ICR0 B3LZY2 B4LY03 A0A182QAD4 A0A0Q9WRP9 A0A1I8PD63 A0A1I8MGW3 W8BB78 W8ARB1 A0A1I8MGW6 T1PHN4 A0A1B0FML0 A0A1B0C9K4 A0A182V4P1 A0A182II23 Q7QDB1 A0A182LFQ7 A0A0A1WVX1 A0A1A9VAY2 A0A1A9W3U2 A0A1Q3G1I7
A0A0L7LRP1 D6W9G4 A0A2J7RC21 A0A1S3DM74 A0A1Y1MY32 A0A1Y1MXZ1 A0A2R7WEP4 A0A023F2X8 V5GHW6 A0A1B6MRI4 T1HMH5 N6TDH0 E0VD76 A0A0T6BHL7 A0A1B6J9F7 A0A2S2R8L8 A0A067RG97 J9JKH9 A0A2H8TNI1 B4NL04 A0A3Q0IMQ1 A0A2S2PEK9 A0A336M2N9 A0A146LZE2 A0A146LK24 A0A146LBD8 B0X4Q2 A0A182QQK7 A0A2M4BK64 A0A182NFM7 A0A182K8I9 A0A182Y4D4 A0A182W088 A0A2M4BIP6 A0A1L8DDY6 A0A182PAB5 F5HKA0 A0A2M4BHC1 A0A182MJ90 B4GMK2 A0A182RNN6 I5APN8 A0A1Y9INC2 W5JEI9 B5DW31 A0A1C7CZB1 A0A1S4HC19 A0A3F2Z0C8 A0A1Y9GLM9 A0A1I8JVP7 A0A0P6J4H9 Q17CS3 A0A0M4ECH5 Q8MQI2 B4QX70 B4HLD0 A0A0R1E318 A2RVG5 A0A0Q9X0P4 B4JG23 B3P0Y9 A0A3B0KIU3 B4KAE2 B4PR32 A0A0Q9XBC6 A0A1W4VVS4 A0A3B0JN53 Q9VEY2 A0A182F7G1 A0A1J1IAV1 A0A1W4VU24 A0A0P9ATM0 A0A182JKT0 A0A1J1ICR0 B3LZY2 B4LY03 A0A182QAD4 A0A0Q9WRP9 A0A1I8PD63 A0A1I8MGW3 W8BB78 W8ARB1 A0A1I8MGW6 T1PHN4 A0A1B0FML0 A0A1B0C9K4 A0A182V4P1 A0A182II23 Q7QDB1 A0A182LFQ7 A0A0A1WVX1 A0A1A9VAY2 A0A1A9W3U2 A0A1Q3G1I7
PDB
6N3I
E-value=0.000358509,
Score=105
Ontologies
GO
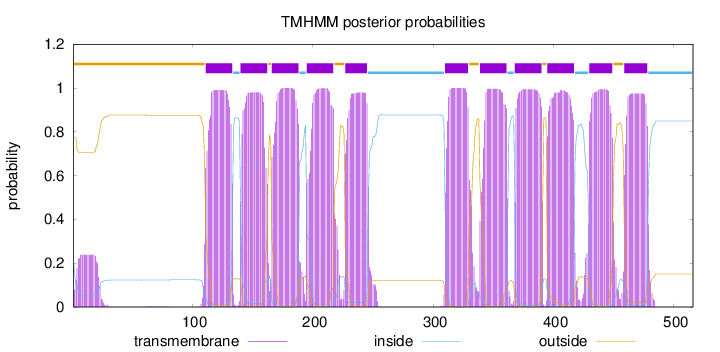
Topology
SignalP
Position: 1 - 25,
Likelihood: 0.555118

Length:
516
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
239.97332
Exp number, first 60 AAs:
4.67298999999999
Total prob of N-in:
0.22586
outside
1 - 110
TMhelix
111 - 133
inside
134 - 139
TMhelix
140 - 162
outside
163 - 165
TMhelix
166 - 188
inside
189 - 194
TMhelix
195 - 217
outside
218 - 226
TMhelix
227 - 245
inside
246 - 309
TMhelix
310 - 329
outside
330 - 338
TMhelix
339 - 361
inside
362 - 367
TMhelix
368 - 390
outside
391 - 394
TMhelix
395 - 417
inside
418 - 429
TMhelix
430 - 449
outside
450 - 458
TMhelix
459 - 478
inside
479 - 516
Population Genetic Test Statistics
Pi
213.110349
Theta
150.155829
Tajima's D
1.458325
CLR
0.372359
CSRT
0.774711264436778
Interpretation
Uncertain
