Gene
KWMTBOMO13696 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011019
Annotation
endothelial-monocyte_activating_polypeptide_II_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.835 Mitochondrial Reliability : 1.221 Nuclear Reliability : 1.541
Sequence
CDS
ATGTTAAAATTGTCATTTGTATTGCGTAGAAATATGTCTCAGTCAGTTACGAAACTTATAAATAATGCAGATATAGCTGAAAAAAGGCTAGCCGAATTACGTAAAAAGCTGGAAGAGGTGAAAAAGTTTAAAATAGAAGAAAAGATCAAACAGCTAATACAAGAAAATAATACTTTGTCGGCGAAGGTAAACGCTGCGAAGGCAGAGTTAATAGGTTTGGAAATATCACACGGAAAGAGGCAGTATGCGATACCTGGTAAAACAGACCTGCTGGAAACAACCAAGATTGTTACAACTATTCAAGATTCCGTTTGTAATGAAAAACCAATTATCGATAAAAAGGAGAAGAAAGTCAAAGATTCTAACAAAAAACCCAAAGATAAACCTAGTGAATCGGCTGATATAGCAGTAGATGTAAGGAAACTAGATTTTCGGATTGGAAAAATTGTTGAGATTAGTAAACACCCTGATGCTGATTCTTTGTATGTGGAAAAAATAGACTGTGGTGAAGAAAATCCACGTACTGTTGTGTCTGGTTTAGTAAATCATGTTCCAATAGATGAAATGCGAGAAAGAATAGTTATGGTACTCTGCAACTTGAAACCAGTTAAGATGAGAGGTGTAACATCTGAAGCCATGGTAATGTGTGCATCATCTGCTGAAAAAGTTGAAGTTTTAATACCTCCACCTGATGCTATACCAGGGGATTTGGTTGTATGTGAAGGATATCCTAGAGAGCCTGAAGCTGTGCTTAATCCCAAAAAGAAAATATTTGAGACTTGTGCACCTGATTTAAAAACAAATAATGAAAAGATAGCCTGTTATAAGGGTAATCCAATAATTGTACCAGGAAAAGGTCCAATAATTGCACCAACTTTAATCAATGTTAATGTAAAATAA
Protein
MLKLSFVLRRNMSQSVTKLINNADIAEKRLAELRKKLEEVKKFKIEEKIKQLIQENNTLSAKVNAAKAELIGLEISHGKRQYAIPGKTDLLETTKIVTTIQDSVCNEKPIIDKKEKKVKDSNKKPKDKPSESADIAVDVRKLDFRIGKIVEISKHPDADSLYVEKIDCGEENPRTVVSGLVNHVPIDEMRERIVMVLCNLKPVKMRGVTSEAMVMCASSAEKVEVLIPPPDAIPGDLVVCEGYPREPEAVLNPKKKIFETCAPDLKTNNEKIACYKGNPIIVPGKGPIIAPTLINVNVK
Summary
Uniprot
Q2F6A4
H9JNB6
A0A2H1VUF5
A0A212ES89
A0A194PN20
A0A2A4K9R2
+ More
A0A1A9UVK1 W8BNV6 A0A0L0C682 A0A034W1W8 A0A0C9QIA5 A0A1B0G734 B4HSB8 D3TLH2 T1PEE1 A0A0J9R9L7 A0A1I8NBA8 A0A1J1HNL1 B3MIA5 B4MIX3 A0A0K8UZN4 A0A1L8DGA3 A1Z7K8 A0A1L8DGD8 A0A0M3QUG6 B0WG59 A0A0R1DMF2 A0A1I8P7P6 B3N840 A0A1W4UXP6 B4KTP9 B4LNP3 A0A1B0DPQ9 Q28Y85 B4GCU9 A0A0R3NU26 B4JVF4 A0A336MH26 A0A3B0JRY8 A0A1B0CJY7 A0A3B0J051 A0A336LVI7 T1GBR2 A0A1Q3F719 A0A0K8TLD8 Q16SR4 Q17AI3 Q7Q8S7 B4NSB8 U5EGV7 U4UMB6 A0A182MMP8 A0A182YA42 J3JVT2 A0A182WCU9 A0A0T6AUI3 D7EJV9 A0A182K4Y2 A0A139W9X2
A0A1A9UVK1 W8BNV6 A0A0L0C682 A0A034W1W8 A0A0C9QIA5 A0A1B0G734 B4HSB8 D3TLH2 T1PEE1 A0A0J9R9L7 A0A1I8NBA8 A0A1J1HNL1 B3MIA5 B4MIX3 A0A0K8UZN4 A0A1L8DGA3 A1Z7K8 A0A1L8DGD8 A0A0M3QUG6 B0WG59 A0A0R1DMF2 A0A1I8P7P6 B3N840 A0A1W4UXP6 B4KTP9 B4LNP3 A0A1B0DPQ9 Q28Y85 B4GCU9 A0A0R3NU26 B4JVF4 A0A336MH26 A0A3B0JRY8 A0A1B0CJY7 A0A3B0J051 A0A336LVI7 T1GBR2 A0A1Q3F719 A0A0K8TLD8 Q16SR4 Q17AI3 Q7Q8S7 B4NSB8 U5EGV7 U4UMB6 A0A182MMP8 A0A182YA42 J3JVT2 A0A182WCU9 A0A0T6AUI3 D7EJV9 A0A182K4Y2 A0A139W9X2
Pubmed
EMBL
DQ311168
ABD36113.1
BABH01019596
ODYU01004489
SOQ44418.1
AGBW02012844
+ More
OWR44355.1 KQ459600 KPI94129.1 NWSH01000020 PCG80706.1 GAMC01003740 JAC02816.1 JRES01000848 KNC27766.1 GAKP01011210 GAKP01011206 GAKP01011204 JAC47748.1 GBYB01014478 JAG84245.1 CCAG010003770 CH480816 EDW47013.1 EZ422274 ADD18550.1 KA646313 AFP60942.1 CM002911 KMY92379.1 CVRI01000010 CRK88980.1 CH902619 EDV35950.2 CH963719 EDW72062.2 GDHF01020296 JAI32018.1 GFDF01008601 JAV05483.1 AE013599 AAF59019.2 GFDF01008556 JAV05528.1 CP012524 ALC40633.1 DS231923 EDS26755.1 CM000157 KRJ98428.1 CH954177 EDV59453.1 CH933808 EDW09632.2 CH940648 EDW60113.2 AJVK01001581 CM000071 EAL26080.2 CH479181 EDW32512.1 KRT02815.1 CH916375 EDV98422.1 UFQT01001192 SSX29415.1 OUUW01000001 SPP73878.1 AJWK01015538 SPP73877.1 UFQT01000226 SSX22056.1 CAQQ02198381 GFDL01011685 JAV23360.1 GDAI01002436 JAI15167.1 CH477668 EAT37505.1 CH477335 EAT43239.1 AAAB01008933 EAA09959.4 CH982072 EDX15496.1 GANO01003286 JAB56585.1 KB632275 ERL91150.1 AXCM01000498 BT127350 AEE62312.1 LJIG01022785 KRT78728.1 DS497720 EFA12886.1 KQ971866 KYB24711.1
OWR44355.1 KQ459600 KPI94129.1 NWSH01000020 PCG80706.1 GAMC01003740 JAC02816.1 JRES01000848 KNC27766.1 GAKP01011210 GAKP01011206 GAKP01011204 JAC47748.1 GBYB01014478 JAG84245.1 CCAG010003770 CH480816 EDW47013.1 EZ422274 ADD18550.1 KA646313 AFP60942.1 CM002911 KMY92379.1 CVRI01000010 CRK88980.1 CH902619 EDV35950.2 CH963719 EDW72062.2 GDHF01020296 JAI32018.1 GFDF01008601 JAV05483.1 AE013599 AAF59019.2 GFDF01008556 JAV05528.1 CP012524 ALC40633.1 DS231923 EDS26755.1 CM000157 KRJ98428.1 CH954177 EDV59453.1 CH933808 EDW09632.2 CH940648 EDW60113.2 AJVK01001581 CM000071 EAL26080.2 CH479181 EDW32512.1 KRT02815.1 CH916375 EDV98422.1 UFQT01001192 SSX29415.1 OUUW01000001 SPP73878.1 AJWK01015538 SPP73877.1 UFQT01000226 SSX22056.1 CAQQ02198381 GFDL01011685 JAV23360.1 GDAI01002436 JAI15167.1 CH477668 EAT37505.1 CH477335 EAT43239.1 AAAB01008933 EAA09959.4 CH982072 EDX15496.1 GANO01003286 JAB56585.1 KB632275 ERL91150.1 AXCM01000498 BT127350 AEE62312.1 LJIG01022785 KRT78728.1 DS497720 EFA12886.1 KQ971866 KYB24711.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000218220
UP000078200
UP000037069
+ More
UP000092444 UP000001292 UP000095301 UP000183832 UP000007801 UP000007798 UP000000803 UP000092553 UP000002320 UP000002282 UP000095300 UP000008711 UP000192221 UP000009192 UP000008792 UP000092462 UP000001819 UP000008744 UP000001070 UP000268350 UP000092461 UP000015102 UP000008820 UP000007062 UP000000304 UP000030742 UP000075883 UP000076408 UP000075920 UP000007266 UP000075881
UP000092444 UP000001292 UP000095301 UP000183832 UP000007801 UP000007798 UP000000803 UP000092553 UP000002320 UP000002282 UP000095300 UP000008711 UP000192221 UP000009192 UP000008792 UP000092462 UP000001819 UP000008744 UP000001070 UP000268350 UP000092461 UP000015102 UP000008820 UP000007062 UP000000304 UP000030742 UP000075883 UP000076408 UP000075920 UP000007266 UP000075881
Pfam
PF01588 tRNA_bind
SUPFAM
SSF50249
SSF50249
ProteinModelPortal
Q2F6A4
H9JNB6
A0A2H1VUF5
A0A212ES89
A0A194PN20
A0A2A4K9R2
+ More
A0A1A9UVK1 W8BNV6 A0A0L0C682 A0A034W1W8 A0A0C9QIA5 A0A1B0G734 B4HSB8 D3TLH2 T1PEE1 A0A0J9R9L7 A0A1I8NBA8 A0A1J1HNL1 B3MIA5 B4MIX3 A0A0K8UZN4 A0A1L8DGA3 A1Z7K8 A0A1L8DGD8 A0A0M3QUG6 B0WG59 A0A0R1DMF2 A0A1I8P7P6 B3N840 A0A1W4UXP6 B4KTP9 B4LNP3 A0A1B0DPQ9 Q28Y85 B4GCU9 A0A0R3NU26 B4JVF4 A0A336MH26 A0A3B0JRY8 A0A1B0CJY7 A0A3B0J051 A0A336LVI7 T1GBR2 A0A1Q3F719 A0A0K8TLD8 Q16SR4 Q17AI3 Q7Q8S7 B4NSB8 U5EGV7 U4UMB6 A0A182MMP8 A0A182YA42 J3JVT2 A0A182WCU9 A0A0T6AUI3 D7EJV9 A0A182K4Y2 A0A139W9X2
A0A1A9UVK1 W8BNV6 A0A0L0C682 A0A034W1W8 A0A0C9QIA5 A0A1B0G734 B4HSB8 D3TLH2 T1PEE1 A0A0J9R9L7 A0A1I8NBA8 A0A1J1HNL1 B3MIA5 B4MIX3 A0A0K8UZN4 A0A1L8DGA3 A1Z7K8 A0A1L8DGD8 A0A0M3QUG6 B0WG59 A0A0R1DMF2 A0A1I8P7P6 B3N840 A0A1W4UXP6 B4KTP9 B4LNP3 A0A1B0DPQ9 Q28Y85 B4GCU9 A0A0R3NU26 B4JVF4 A0A336MH26 A0A3B0JRY8 A0A1B0CJY7 A0A3B0J051 A0A336LVI7 T1GBR2 A0A1Q3F719 A0A0K8TLD8 Q16SR4 Q17AI3 Q7Q8S7 B4NSB8 U5EGV7 U4UMB6 A0A182MMP8 A0A182YA42 J3JVT2 A0A182WCU9 A0A0T6AUI3 D7EJV9 A0A182K4Y2 A0A139W9X2
PDB
4R3Z
E-value=1.61207e-49,
Score=494
Ontologies
GO
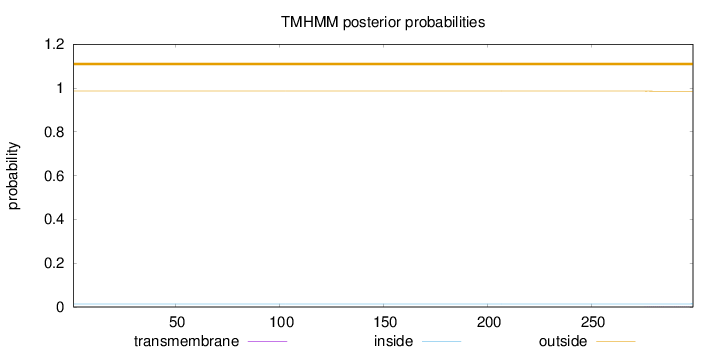
Topology
Length:
299
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01948
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01381
outside
1 - 299
Population Genetic Test Statistics
Pi
200.787256
Theta
200.111472
Tajima's D
0.019746
CLR
0.552457
CSRT
0.370331483425829
Interpretation
Uncertain
