Gene
KWMTBOMO13692
Pre Gene Modal
BGIBMGA011022
Annotation
PREDICTED:_bursicon_isoform_X1_[Bombyx_mori]
Full name
Bursicon
Alternative Name
Bursicon subunit alpha
Cuticle-tanning hormone
Single-chain bursicon
Cuticle-tanning hormone
Single-chain bursicon
Location in the cell
Extracellular Reliability : 4.008
Sequence
CDS
ATGTCCGTATTGAATACTTTTTTAGTCATAGTGGCTTTAATCTTATGTTACGTAAATGATTTCCCTGTTACTGGGCATGAAGTTCAACTACCTCCAGGCCAGGAATGTCAAATGACCGCAGTCATTCATGTTTTAAAACACCGAGGATGTAAACCTAAAGCAATTCCGTCGTTTGCTTGCATTGGAAAATGCACAAGCTACGTTCAGGTATCGGGTAGTAAAATATGGCAAATGGAGCGTACCTGCAACTGCTGTCAAGAATCAGGAGAAAGAGAAGCTACAGTTGTCTTGTTTTGTCCGGATGCTCAAAATGAAGAAAAGCGATTCAGAAAGGTTTCAACGAAAGCTCCGCTTCAATGCATGTGTCGTCCTTGCGGAAGTATAGAAGAGAGTTCCATTATTCCTCAGGAAGTTGCTGGATACTCCGAAGAAGGACCGCTGTATAACCATTTCAGGAAGTCTCTTTAA
Protein
MSVLNTFLVIVALILCYVNDFPVTGHEVQLPPGQECQMTAVIHVLKHRGCKPKAIPSFACIGKCTSYVQVSGSKIWQMERTCNCCQESGEREATVVLFCPDAQNEEKRFRKVSTKAPLQCMCRPCGSIEESSIIPQEVAGYSEEGPLYNHFRKSL
Summary
Description
Final heterodimeric neurohormone released at the end of the molting cycle, involved in the sclerotization (tanning) of the insect cuticle, melanization and wing spreading.
Final heterodimeric neurohormone released at the end of the molting cycle, involved in the sclerotization (tanning) of the insect cuticle, melanization and wing spreading. Heterodimer specifically activates the G protein-coupled receptor rk.
Final heterodimeric neurohormone released at the end of the molting cycle, involved in the sclerotization (tanning) of the insect cuticle, melanization and wing spreading. Heterodimer specifically activates the G protein-coupled receptor rk.
Subunit
Heterodimer of burs and pburs.
Heterodimer of Burs and Pburs.
Heterodimer of Burs and Pburs.
Miscellaneous
The bursicon gene is encoded by two loci: burs124 contains exons 1, 2 and 4, and burs3 contains exon 3. Exon 3 is trans-spliced into position in the mature transcript. This unusual gene arrangement has existed for at least the 150 million years (MY) of mosquito evolution since the split of the culicid and anophelid family lineages, but is younger than the 250 MY split of the dipteran suborders Nematocera and Brachycera (containing the Culicidea and Drosophilidae, respectively).
The bursicon gene is encoded by two loci: burs124 on chromosome arm 2L contains exons 1, 2 and 4, and burs3 on arm 2R contains exon 3. Exon 3 is trans-spliced into position in the mature transcript. This unusual gene arrangement has existed for at least the 150 million years (MY) of mosquito evolution since the split of the culicid and anophelid family lineages, but is younger than the 250 MY split of the dipteran suborders Nematocera and Brachycera (containing the Culicidea and Drosophilidae, respectively).
The bursicon gene is encoded by two loci: burs124 on chromosome arm 2L contains exons 1, 2 and 4, and burs3 on arm 2R contains exon 3. Exon 3 is trans-spliced into position in the mature transcript. This unusual gene arrangement has existed for at least the 150 million years (MY) of mosquito evolution since the split of the culicid and anophelid family lineages, but is younger than the 250 MY split of the dipteran suborders Nematocera and Brachycera (containing the Culicidea and Drosophilidae, respectively).
Keywords
Complete proteome
Disulfide bond
Hormone
Reference proteome
Secreted
Signal
Feature
chain Bursicon
Uniprot
Q566B1
Q4FCM6
A0A2H1WNT1
A0A385H8U1
A0A212ES77
A0A0S1U1M9
+ More
A0A2H1VUC7 A0A2A4K923 A0A3S2TSN9 W8RXF2 A0A0C5BEX6 A0A194PN16 A0A194QN72 V5NFD5 A0A0L7K2C7 A0A067R3X7 A0A2P8YCI2 A0A143WDY5 A0A2J7PJC7 W8C890 A0A034V006 A0A1B6GB49 A0A0A1WZR0 A0A1B0GDZ7 A0A1B0AHG7 A0A1A9Y4J5 A0A1B0BBC4 B4K981 B4JHI8 B4M141 B4HM41 A0APK7 C0MK54 Q9VD83 B4PKV0 A0A1I8M3F5 A0A1W4W143 A0A0H3YID2 B4NIZ9 B3NZY3 B3LVH1 Q29AR9 B4G615 A0A1B0CEW8 A0A3B0K4N7 U3U8R1 A0A0M4F2F2 A0A1I8NY16 A0A345BED3 A0RZD3 A0A1W4W3Y0 E0VXD8 B5FX32 A0A1E1G7Q6 A3RL96 A0A3Q8KJL9 A0A336KB63 A0A336KZK4 P85316 A0A0K8TGB5 Q66Q82 P85315 A0A1S3CW54 A0A2R4SUG4 J9JXY0 A0A2S2Q949 A0A2H8TSR9 A0A2S2PBD6 A0A154NY08 A0A151IK56 A0A151WRJ9 A0A0N0U3I1 A2VB89 A0A310SSL9 E2A3Q3 A0A195B087 A0A195FC54 A0A151JB20 A0A3L8DDW9 A0A1A9VM32 A0A2A3E9R4 E9J7U4 A0A026WU74 E2BTT5 A0A158NKI9 I3VN95 B0LI27 E9FYK9 A0A164WQ02 A0A0P5FVZ6 A0A0P5IV69 A0A3R7MBC0 A0A0G3EW09 F1AQ58 T1JJS2 A0A0K2UN46 D9I4C3 A0A2P2I1H7 A0A2P6KMT9
A0A2H1VUC7 A0A2A4K923 A0A3S2TSN9 W8RXF2 A0A0C5BEX6 A0A194PN16 A0A194QN72 V5NFD5 A0A0L7K2C7 A0A067R3X7 A0A2P8YCI2 A0A143WDY5 A0A2J7PJC7 W8C890 A0A034V006 A0A1B6GB49 A0A0A1WZR0 A0A1B0GDZ7 A0A1B0AHG7 A0A1A9Y4J5 A0A1B0BBC4 B4K981 B4JHI8 B4M141 B4HM41 A0APK7 C0MK54 Q9VD83 B4PKV0 A0A1I8M3F5 A0A1W4W143 A0A0H3YID2 B4NIZ9 B3NZY3 B3LVH1 Q29AR9 B4G615 A0A1B0CEW8 A0A3B0K4N7 U3U8R1 A0A0M4F2F2 A0A1I8NY16 A0A345BED3 A0RZD3 A0A1W4W3Y0 E0VXD8 B5FX32 A0A1E1G7Q6 A3RL96 A0A3Q8KJL9 A0A336KB63 A0A336KZK4 P85316 A0A0K8TGB5 Q66Q82 P85315 A0A1S3CW54 A0A2R4SUG4 J9JXY0 A0A2S2Q949 A0A2H8TSR9 A0A2S2PBD6 A0A154NY08 A0A151IK56 A0A151WRJ9 A0A0N0U3I1 A2VB89 A0A310SSL9 E2A3Q3 A0A195B087 A0A195FC54 A0A151JB20 A0A3L8DDW9 A0A1A9VM32 A0A2A3E9R4 E9J7U4 A0A026WU74 E2BTT5 A0A158NKI9 I3VN95 B0LI27 E9FYK9 A0A164WQ02 A0A0P5FVZ6 A0A0P5IV69 A0A3R7MBC0 A0A0G3EW09 F1AQ58 T1JJS2 A0A0K2UN46 D9I4C3 A0A2P2I1H7 A0A2P6KMT9
Pubmed
15141943
15811337
30171635
22118469
25640113
26354079
+ More
26227816 24845553 29403074 26706852 24495485 25348373 25830018 17994087 16951084 19126864 15242619 10731132 12537572 15703293 17550304 25315136 26036749 18057021 15632085 23932938 18362917 19820115 20566863 30467353 17435221 17510324 12364791 17275819 20798317 30249741 21282665 24508170 21347285 18221939 21292972 20691691
26227816 24845553 29403074 26706852 24495485 25348373 25830018 17994087 16951084 19126864 15242619 10731132 12537572 15703293 17550304 25315136 26036749 18057021 15632085 23932938 18362917 19820115 20566863 30467353 17435221 17510324 12364791 17275819 20798317 30249741 21282665 24508170 21347285 18221939 21292972 20691691
EMBL
BAAB01143969
BN000691
CAH89262.2
DQ094149
AAZ04374.1
ODYU01009965
+ More
SOQ54731.1 MH028069 AXY04238.1 AGBW02012844 OWR44350.1 KT005955 ALM30306.1 ODYU01004489 SOQ44423.1 NWSH01000020 PCG80721.1 RSAL01000008 RVE54039.1 KJ002445 AHM02472.1 KJ783481 AJM76770.1 KQ459600 KPI94124.1 KQ461195 KPJ06804.1 KF498645 AHA86938.1 JTDY01014723 JTDY01000043 KOB52000.1 KOB79213.1 KK852914 KDR13886.1 PYGN01000705 PSN41953.1 LN901328 CUT08823.1 NEVH01024949 PNF16418.1 GAMC01000052 JAC06504.1 GAKP01023486 JAC35472.1 GECZ01010127 JAS59642.1 GBXI01009995 JAD04297.1 CCAG010012076 JXJN01011407 CH933806 EDW14494.1 CH916369 EDV92815.1 CH940650 EDW67452.1 CH480815 EDW43089.1 AM294595 CM000364 CAL26578.1 EDX13824.1 BT122042 BT132842 FM245726 FM245727 FM245728 FM245729 FM245730 FM245731 FM245732 FM245733 FM245734 FM245735 FM245736 ADD31622.1 AEU10774.1 CAR93652.1 CAR93653.1 CAR93654.1 CAR93655.1 CAR93656.1 CAR93657.1 CAR93658.1 CAR93659.1 CAR93660.1 CAR93661.1 CAR93662.1 AY672905 AY672906 AJ862523 AM294583 AM294584 AM294585 AM294586 AM294587 AM294588 AM294589 AM294590 AM294591 AM294592 AM294593 AM294594 AE014297 AAF55915.1 AAT72327.1 CM000160 EDW97899.2 KP895534 AKN21233.1 CH964272 EDW84901.1 CH954462 CH954181 EDV45165.1 EDV49981.1 CH902617 EDV42541.1 CM000070 EAL27280.1 CH479179 EDW23769.1 AJWK01009369 OUUW01000007 SPP82960.1 AB817240 BAO00937.1 CP012526 ALC45612.1 MF765465 AXF48185.1 DQ138189 KQ971342 ABA03053.1 EFA04634.1 DS235830 EEB18044.1 DQ156996 ABA40402.1 LC146489 BAV78793.1 EF424614 ABO20870.1 MH971162 AZC86174.1 UFQS01000224 UFQT01000224 SSX01647.1 SSX22027.1 UFQS01001316 UFQT01001316 SSX10316.1 SSX30004.1 CH477322 CH477412 GBRD01001302 JAG64519.1 AY735442 AY735443 AAAB01008859 AAAB01008900 MF083566 AVZ65957.1 ABLF02027799 GGMS01005041 MBY74244.1 GFXV01005471 MBW17276.1 GGMR01014141 MBY26760.1 KQ434772 KZC03908.1 KQ977294 KYN03905.1 KQ982805 KYQ50474.1 KQ435881 KOX69832.1 AM420631 AADG06006208 BN000692 CAH89263.2 CAM06631.1 KQ759843 OAD62600.1 GL436457 EFN71945.1 KQ976692 KYM77871.1 KQ981693 KYN37782.1 KQ979182 KYN22332.1 QOIP01000009 RLU18680.1 KZ288311 PBC28465.1 GL768630 EFZ11126.1 KK107119 EZA58629.1 GL450531 EFN80834.1 ADTU01018857 ADTU01018858 JQ864194 AFK81932.1 EU139431 ABX55998.1 GL732527 EFX87546.1 LRGB01001151 KZS13469.1 GDIQ01271102 JAJ80622.1 GDIQ01208625 JAK43100.1 QCYY01001562 ROT77167.1 KP191597 KT072701 AKJ74864.1 ALO17551.1 GU937434 ADY80040.1 JH431954 HACA01022343 CDW39704.1 HM113369 ADI86242.1 IACF01002186 LAB67849.1 MWRG01008500 PRD27627.1
SOQ54731.1 MH028069 AXY04238.1 AGBW02012844 OWR44350.1 KT005955 ALM30306.1 ODYU01004489 SOQ44423.1 NWSH01000020 PCG80721.1 RSAL01000008 RVE54039.1 KJ002445 AHM02472.1 KJ783481 AJM76770.1 KQ459600 KPI94124.1 KQ461195 KPJ06804.1 KF498645 AHA86938.1 JTDY01014723 JTDY01000043 KOB52000.1 KOB79213.1 KK852914 KDR13886.1 PYGN01000705 PSN41953.1 LN901328 CUT08823.1 NEVH01024949 PNF16418.1 GAMC01000052 JAC06504.1 GAKP01023486 JAC35472.1 GECZ01010127 JAS59642.1 GBXI01009995 JAD04297.1 CCAG010012076 JXJN01011407 CH933806 EDW14494.1 CH916369 EDV92815.1 CH940650 EDW67452.1 CH480815 EDW43089.1 AM294595 CM000364 CAL26578.1 EDX13824.1 BT122042 BT132842 FM245726 FM245727 FM245728 FM245729 FM245730 FM245731 FM245732 FM245733 FM245734 FM245735 FM245736 ADD31622.1 AEU10774.1 CAR93652.1 CAR93653.1 CAR93654.1 CAR93655.1 CAR93656.1 CAR93657.1 CAR93658.1 CAR93659.1 CAR93660.1 CAR93661.1 CAR93662.1 AY672905 AY672906 AJ862523 AM294583 AM294584 AM294585 AM294586 AM294587 AM294588 AM294589 AM294590 AM294591 AM294592 AM294593 AM294594 AE014297 AAF55915.1 AAT72327.1 CM000160 EDW97899.2 KP895534 AKN21233.1 CH964272 EDW84901.1 CH954462 CH954181 EDV45165.1 EDV49981.1 CH902617 EDV42541.1 CM000070 EAL27280.1 CH479179 EDW23769.1 AJWK01009369 OUUW01000007 SPP82960.1 AB817240 BAO00937.1 CP012526 ALC45612.1 MF765465 AXF48185.1 DQ138189 KQ971342 ABA03053.1 EFA04634.1 DS235830 EEB18044.1 DQ156996 ABA40402.1 LC146489 BAV78793.1 EF424614 ABO20870.1 MH971162 AZC86174.1 UFQS01000224 UFQT01000224 SSX01647.1 SSX22027.1 UFQS01001316 UFQT01001316 SSX10316.1 SSX30004.1 CH477322 CH477412 GBRD01001302 JAG64519.1 AY735442 AY735443 AAAB01008859 AAAB01008900 MF083566 AVZ65957.1 ABLF02027799 GGMS01005041 MBY74244.1 GFXV01005471 MBW17276.1 GGMR01014141 MBY26760.1 KQ434772 KZC03908.1 KQ977294 KYN03905.1 KQ982805 KYQ50474.1 KQ435881 KOX69832.1 AM420631 AADG06006208 BN000692 CAH89263.2 CAM06631.1 KQ759843 OAD62600.1 GL436457 EFN71945.1 KQ976692 KYM77871.1 KQ981693 KYN37782.1 KQ979182 KYN22332.1 QOIP01000009 RLU18680.1 KZ288311 PBC28465.1 GL768630 EFZ11126.1 KK107119 EZA58629.1 GL450531 EFN80834.1 ADTU01018857 ADTU01018858 JQ864194 AFK81932.1 EU139431 ABX55998.1 GL732527 EFX87546.1 LRGB01001151 KZS13469.1 GDIQ01271102 JAJ80622.1 GDIQ01208625 JAK43100.1 QCYY01001562 ROT77167.1 KP191597 KT072701 AKJ74864.1 ALO17551.1 GU937434 ADY80040.1 JH431954 HACA01022343 CDW39704.1 HM113369 ADI86242.1 IACF01002186 LAB67849.1 MWRG01008500 PRD27627.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000283053
UP000053268
UP000053240
+ More
UP000037510 UP000027135 UP000245037 UP000235965 UP000092444 UP000092445 UP000092443 UP000092460 UP000009192 UP000001070 UP000008792 UP000001292 UP000000304 UP000000803 UP000002282 UP000095301 UP000192221 UP000007798 UP000008711 UP000007801 UP000001819 UP000008744 UP000092461 UP000268350 UP000092553 UP000095300 UP000007266 UP000192223 UP000009046 UP000008820 UP000007062 UP000079169 UP000007819 UP000076502 UP000078542 UP000075809 UP000053105 UP000005203 UP000000311 UP000078540 UP000078541 UP000078492 UP000279307 UP000078200 UP000242457 UP000053097 UP000008237 UP000005205 UP000000305 UP000076858 UP000283509
UP000037510 UP000027135 UP000245037 UP000235965 UP000092444 UP000092445 UP000092443 UP000092460 UP000009192 UP000001070 UP000008792 UP000001292 UP000000304 UP000000803 UP000002282 UP000095301 UP000192221 UP000007798 UP000008711 UP000007801 UP000001819 UP000008744 UP000092461 UP000268350 UP000092553 UP000095300 UP000007266 UP000192223 UP000009046 UP000008820 UP000007062 UP000079169 UP000007819 UP000076502 UP000078542 UP000075809 UP000053105 UP000005203 UP000000311 UP000078540 UP000078541 UP000078492 UP000279307 UP000078200 UP000242457 UP000053097 UP000008237 UP000005205 UP000000305 UP000076858 UP000283509
Interpro
Gene 3D
CDD
ProteinModelPortal
Q566B1
Q4FCM6
A0A2H1WNT1
A0A385H8U1
A0A212ES77
A0A0S1U1M9
+ More
A0A2H1VUC7 A0A2A4K923 A0A3S2TSN9 W8RXF2 A0A0C5BEX6 A0A194PN16 A0A194QN72 V5NFD5 A0A0L7K2C7 A0A067R3X7 A0A2P8YCI2 A0A143WDY5 A0A2J7PJC7 W8C890 A0A034V006 A0A1B6GB49 A0A0A1WZR0 A0A1B0GDZ7 A0A1B0AHG7 A0A1A9Y4J5 A0A1B0BBC4 B4K981 B4JHI8 B4M141 B4HM41 A0APK7 C0MK54 Q9VD83 B4PKV0 A0A1I8M3F5 A0A1W4W143 A0A0H3YID2 B4NIZ9 B3NZY3 B3LVH1 Q29AR9 B4G615 A0A1B0CEW8 A0A3B0K4N7 U3U8R1 A0A0M4F2F2 A0A1I8NY16 A0A345BED3 A0RZD3 A0A1W4W3Y0 E0VXD8 B5FX32 A0A1E1G7Q6 A3RL96 A0A3Q8KJL9 A0A336KB63 A0A336KZK4 P85316 A0A0K8TGB5 Q66Q82 P85315 A0A1S3CW54 A0A2R4SUG4 J9JXY0 A0A2S2Q949 A0A2H8TSR9 A0A2S2PBD6 A0A154NY08 A0A151IK56 A0A151WRJ9 A0A0N0U3I1 A2VB89 A0A310SSL9 E2A3Q3 A0A195B087 A0A195FC54 A0A151JB20 A0A3L8DDW9 A0A1A9VM32 A0A2A3E9R4 E9J7U4 A0A026WU74 E2BTT5 A0A158NKI9 I3VN95 B0LI27 E9FYK9 A0A164WQ02 A0A0P5FVZ6 A0A0P5IV69 A0A3R7MBC0 A0A0G3EW09 F1AQ58 T1JJS2 A0A0K2UN46 D9I4C3 A0A2P2I1H7 A0A2P6KMT9
A0A2H1VUC7 A0A2A4K923 A0A3S2TSN9 W8RXF2 A0A0C5BEX6 A0A194PN16 A0A194QN72 V5NFD5 A0A0L7K2C7 A0A067R3X7 A0A2P8YCI2 A0A143WDY5 A0A2J7PJC7 W8C890 A0A034V006 A0A1B6GB49 A0A0A1WZR0 A0A1B0GDZ7 A0A1B0AHG7 A0A1A9Y4J5 A0A1B0BBC4 B4K981 B4JHI8 B4M141 B4HM41 A0APK7 C0MK54 Q9VD83 B4PKV0 A0A1I8M3F5 A0A1W4W143 A0A0H3YID2 B4NIZ9 B3NZY3 B3LVH1 Q29AR9 B4G615 A0A1B0CEW8 A0A3B0K4N7 U3U8R1 A0A0M4F2F2 A0A1I8NY16 A0A345BED3 A0RZD3 A0A1W4W3Y0 E0VXD8 B5FX32 A0A1E1G7Q6 A3RL96 A0A3Q8KJL9 A0A336KB63 A0A336KZK4 P85316 A0A0K8TGB5 Q66Q82 P85315 A0A1S3CW54 A0A2R4SUG4 J9JXY0 A0A2S2Q949 A0A2H8TSR9 A0A2S2PBD6 A0A154NY08 A0A151IK56 A0A151WRJ9 A0A0N0U3I1 A2VB89 A0A310SSL9 E2A3Q3 A0A195B087 A0A195FC54 A0A151JB20 A0A3L8DDW9 A0A1A9VM32 A0A2A3E9R4 E9J7U4 A0A026WU74 E2BTT5 A0A158NKI9 I3VN95 B0LI27 E9FYK9 A0A164WQ02 A0A0P5FVZ6 A0A0P5IV69 A0A3R7MBC0 A0A0G3EW09 F1AQ58 T1JJS2 A0A0K2UN46 D9I4C3 A0A2P2I1H7 A0A2P6KMT9
PDB
4YU8
E-value=0.000920618,
Score=95
Ontologies
GO
PANTHER
Topology
Subcellular location
Secreted
SignalP
Position: 1 - 25,
Likelihood: 0.993676
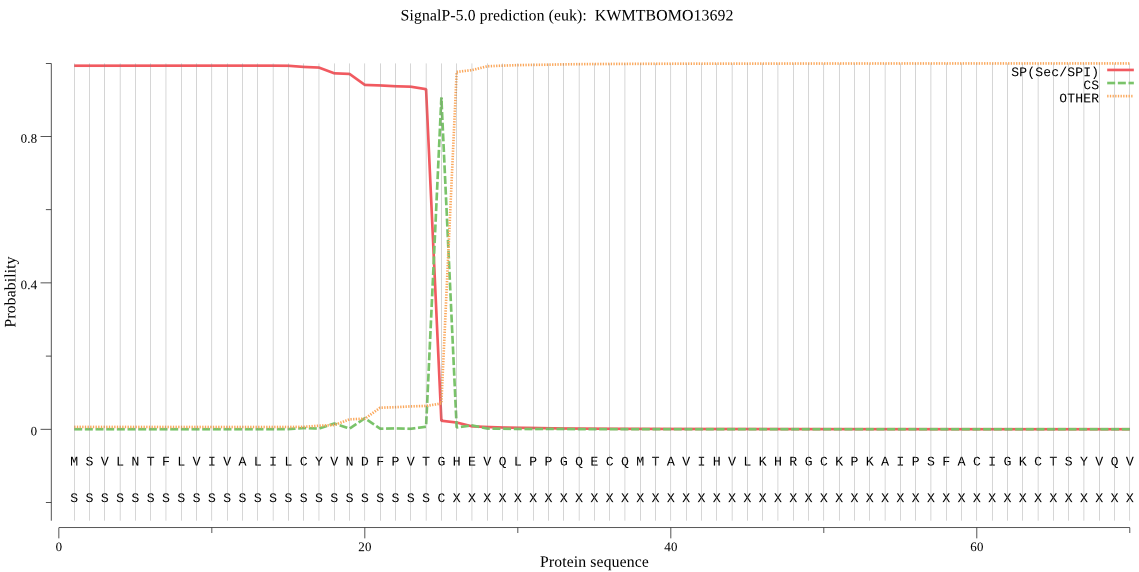
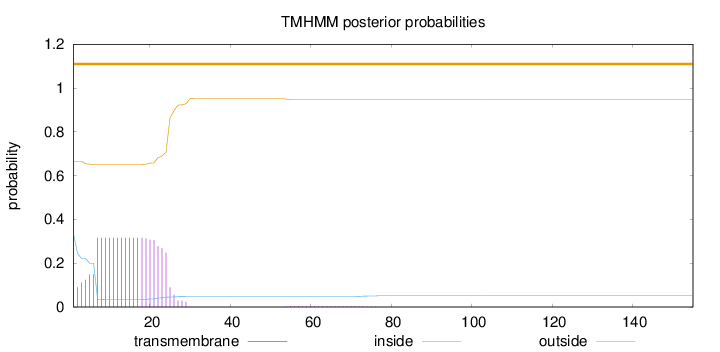
Length:
155
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
6.44567
Exp number, first 60 AAs:
6.39357
Total prob of N-in:
0.33462
outside
1 - 155
Population Genetic Test Statistics
Pi
189.572945
Theta
135.894926
Tajima's D
1.934077
CLR
0
CSRT
0.873106344682766
Interpretation
Uncertain
