Gene
KWMTBOMO13691
Pre Gene Modal
BGIBMGA011022
Annotation
PREDICTED:_lysine-specific_histone_demethylase_1A_[Bombyx_mori]
Full name
Probable lysine-specific histone demethylase 1
+ More
Lysine-specific histone demethylase
Lysine-specific histone demethylase
Location in the cell
Nuclear Reliability : 3.665
Sequence
CDS
ATGAGTCGCCGTAAGCGTGCTAAGGTAGAAAATCGTGAATTAGATGAGAAGCTCAAACAAGAAGATTCTGAATCCGATCAATCCGAGAGAAGTAAAGTCACATCTCCGAGTCCTACGTCAGTGGCAGGTCGCAGAGGGTCGTCCTCAAGTCGTACAGCCGACAACAGCAAGGAGCCTAAAAAAGAAGAAAAAGCTTCTCCAGAGGATCCAAAGAGTGACGATGTTGAAGAAATTAACTATAAAGAAATGCTTAATAGTCTTGAAGGAGCAGCATTTCAGTCGCGATTACCATTTGATAAGATGTCGTCACTTGAAGCAGAATGCTTTTCTGATTTACAAGGCCCTTCGAATTGCGCATTTATTCAAATTCGAAATCGGATTTTAAAAATGTGGTTTGCAGACCCTAAGAAACAGTTGACTCAAGAACAGGCTGTAAAAAAAATGGAGCCTCCCTATAATGGAGATCCAGCATTAATAATGAGAACTCATGCCTTTCTGGAGAGACATGGTTTTATCAACTATGGAATATATGAGCGCGTAACACCAATCCCAAGCCCTCCAAAAGGAAAACAACGCCCAAAAATTATAATTATTGGAGCAGGAGTGTCCGGCTTGGCAGCTGCTCGCCAACTTCAGTCCTTTGGCTGTGAAGTCGTGGTTTTAGAAGGAAGAGACAGAGTCGGAGGCCGAATAGTGACCTACCGGAAAGGACCCTATGTAGCTGACCTAGGTGCTATGGTAGTAACTGGACTAGGAGGTAACCCTGTGACAACGTTGTCAGTGCAAATGAACATGGAGCTTCATAAAATTAAACAAAAATGTCCCCTTTATGAAGCTAATGGGAACCAAGTACCTAAACATAAGGATGAAATGGTGGAACGCGAATTCAATCGTCTCCTAGATGCCACATCTTATCTGTCTCACCAAGTGGATTTCAATTATTATAACGACACGCCTGTCTCACTGGGTCAAGCTTTAGAGTGGGTCATCAAGCTGCAACAAAAAAATGTTAAACATAAACAGATACAGCATCTGAAAGCTCTCACAACGTTGCAAGAGAAAATGAAATCAAACATAAATAAAATGACCGACCTGCAGGATCTCCTCAAGTCCCTGAAATCGGAGAAGGAGGCTTTGCTCGCCGAGAAAGCGACCGGTAATACCGGGGATGCTAATATACAGCAAGAGTTCAATTTGCGGCGACTCAATAGGGAAATGAAACATCTTTGTAACGAATACGAGAATTTGGTCACTCAGAACTCGACTATTGAAGAGAAACTGACACATCTGGAAGCTAATCCGCCGAGTTCCGTGTATCTGTCGGTCCGGGATCGTCAGATATTGGATTGGCACTTTGCGAATTTGGAATTTGCGAACGCAACGCCTCTCGGGAACTTATCGCTGAAGCACTGGGACCAGGACGACGATTTCGAGTTCACCGGAAATCATTTAACAGAGGTAAAACATTACAAAACTTTATAA
Protein
MSRRKRAKVENRELDEKLKQEDSESDQSERSKVTSPSPTSVAGRRGSSSSRTADNSKEPKKEEKASPEDPKSDDVEEINYKEMLNSLEGAAFQSRLPFDKMSSLEAECFSDLQGPSNCAFIQIRNRILKMWFADPKKQLTQEQAVKKMEPPYNGDPALIMRTHAFLERHGFINYGIYERVTPIPSPPKGKQRPKIIIIGAGVSGLAAARQLQSFGCEVVVLEGRDRVGGRIVTYRKGPYVADLGAMVVTGLGGNPVTTLSVQMNMELHKIKQKCPLYEANGNQVPKHKDEMVEREFNRLLDATSYLSHQVDFNYYNDTPVSLGQALEWVIKLQQKNVKHKQIQHLKALTTLQEKMKSNINKMTDLQDLLKSLKSEKEALLAEKATGNTGDANIQQEFNLRRLNREMKHLCNEYENLVTQNSTIEEKLTHLEANPPSSVYLSVRDRQILDWHFANLEFANATPLGNLSLKHWDQDDDFEFTGNHLTEVKHYKTL
Summary
Description
Probable histone demethylase that specifically demethylates 'Lys-4' of histone H3, a specific tag for epigenetic transcriptional activation, thereby acting as a corepressor. Acts by oxidizing the substrate by FAD to generate the corresponding imine that is subsequently hydrolyzed. Demethylates both mono- and di-methylated 'Lys-4' of histone H3.
Histone demethylase that demethylates both 'Lys-4' (H3K4me) and 'Lys-9' (H3K9me) of histone H3, thereby acting as a coactivator or a corepressor, depending on the context. Acts by oxidizing the substrate by FAD to generate the corresponding imine that is subsequently hydrolyzed. Acts as a corepressor by mediating demethylation of H3K4me, a specific tag for epigenetic transcriptional activation. Demethylates both mono- (H3K4me1) and di-methylated (H3K4me2) H3K4me.
Histone demethylase that demethylates both 'Lys-4' (H3K4me) and 'Lys-9' (H3K9me) of histone H3, thereby acting as a coactivator or a corepressor, depending on the context. Acts by oxidizing the substrate by FAD to generate the corresponding imine that is subsequently hydrolyzed. Acts as a corepressor by mediating demethylation of H3K4me, a specific tag for epigenetic transcriptional activation. Demethylates both mono- (H3K4me1) and di-methylated (H3K4me2) H3K4me.
Cofactor
FAD
Similarity
Belongs to the flavin monoamine oxidase family.
Uniprot
H9JNB9
A0A2H1VW42
A0A2A4KAF8
A0A3S2M9H5
A0A194PMA9
A0A212ESA4
+ More
A0A0P6G371 A0A0N8ECW4 A0A087UR95 A0A067RQM1 A0A232FAV7 A0A2J7QTA8 A0A0P5EG14 E9FRK1 A0A2L2Y9V1 A0A0P5BDK1 A0A0N8ADD8 A0A0P5MZV8 A0A0P5SJV5 A0A0P5HAY8 A0A0P5TRG1 A0A0P6D054 B7PX42 A0A0P5HDB5 A0A0P5CQS7 A0A0P5GN72 A0A0P5Y0G1 A0A131XSP8 A0A0N7ZPJ6 A0A0P4YCL5 A0A0P5XXE8 A0A2P6LBY2 A0A0P6GBD1 A0A0N8CZ14 A0A0P5WGT4 A0A0N8DI44 A0A0N8CUD8 A0A1Z5L217 E0VDA2 A0A0P5LKM9 A0A2A3EK20 A0A093J4S8 A0A224Z3J6 A0A0P6I601 A0A0P5AR78 A0A0P5VYP3 A0A0P4Y249 A0A293LJU0 A0A1Z5L5L5 A0A146MDQ3 A0A139WNM3 A0A224YR83 A0A3L8D8C0 A0A091THR5 A0A093FL99 A0A0N0BIS0 A0A093QHP7 E2B2H0 A0A023GP06 A0A0L7R4L4 A0A2R5L6N5 A0A067RCK0 A0A154PCN1 A0A2P8XW97 A0A091P5C4 A0A1V4JCQ2 A0A2D4I895 A0A091I3T2 E2AM99 A0A091H8L9 A0A091JUT6 A0A093QUK5 A0A091LCQ0 A0A091G9L6 A0A158NQ99 R0JCF4 A0A293N5M7 A0A091W1H8 I3JBC0 A0A093G052 A0A2R8Q8P2 A0A093G3F3 A0A091EMG5 A0A091UL68 A0A091JPR1 A0A0A0B0G1 A0A087RGC1 A0A3B4GPL8 A0A0C9RC82 A0A1V4JCG5 A0A093KBC1 A0A093PTF6 A0A088AAG3 A0A226NQN8 A0A099ZEK4 A0A3Q2V816 A0A3L8S3J3 A0A218UY33 A0A3M0IRN7 U3IUI0
A0A0P6G371 A0A0N8ECW4 A0A087UR95 A0A067RQM1 A0A232FAV7 A0A2J7QTA8 A0A0P5EG14 E9FRK1 A0A2L2Y9V1 A0A0P5BDK1 A0A0N8ADD8 A0A0P5MZV8 A0A0P5SJV5 A0A0P5HAY8 A0A0P5TRG1 A0A0P6D054 B7PX42 A0A0P5HDB5 A0A0P5CQS7 A0A0P5GN72 A0A0P5Y0G1 A0A131XSP8 A0A0N7ZPJ6 A0A0P4YCL5 A0A0P5XXE8 A0A2P6LBY2 A0A0P6GBD1 A0A0N8CZ14 A0A0P5WGT4 A0A0N8DI44 A0A0N8CUD8 A0A1Z5L217 E0VDA2 A0A0P5LKM9 A0A2A3EK20 A0A093J4S8 A0A224Z3J6 A0A0P6I601 A0A0P5AR78 A0A0P5VYP3 A0A0P4Y249 A0A293LJU0 A0A1Z5L5L5 A0A146MDQ3 A0A139WNM3 A0A224YR83 A0A3L8D8C0 A0A091THR5 A0A093FL99 A0A0N0BIS0 A0A093QHP7 E2B2H0 A0A023GP06 A0A0L7R4L4 A0A2R5L6N5 A0A067RCK0 A0A154PCN1 A0A2P8XW97 A0A091P5C4 A0A1V4JCQ2 A0A2D4I895 A0A091I3T2 E2AM99 A0A091H8L9 A0A091JUT6 A0A093QUK5 A0A091LCQ0 A0A091G9L6 A0A158NQ99 R0JCF4 A0A293N5M7 A0A091W1H8 I3JBC0 A0A093G052 A0A2R8Q8P2 A0A093G3F3 A0A091EMG5 A0A091UL68 A0A091JPR1 A0A0A0B0G1 A0A087RGC1 A0A3B4GPL8 A0A0C9RC82 A0A1V4JCG5 A0A093KBC1 A0A093PTF6 A0A088AAG3 A0A226NQN8 A0A099ZEK4 A0A3Q2V816 A0A3L8S3J3 A0A218UY33 A0A3M0IRN7 U3IUI0
EC Number
1.-.-.-
Pubmed
EMBL
BABH01019588
BABH01019589
ODYU01004489
SOQ44424.1
NWSH01000020
PCG80720.1
+ More
RSAL01000008 RVE54040.1 KQ459600 KPI94123.1 AGBW02012844 OWR44349.1 GDIQ01039155 JAN55582.1 GDIQ01039156 JAN55581.1 KK121162 KFM79884.1 KK852482 KDR22930.1 NNAY01000500 OXU27976.1 NEVH01011197 PNF31816.1 GDIP01143631 JAJ79771.1 GL732523 EFX90178.1 IAAA01016543 LAA04952.1 GDIP01186028 JAJ37374.1 GDIP01158433 JAJ64969.1 GDIQ01171207 JAK80518.1 GDIP01142541 JAL61173.1 GDIQ01229184 JAK22541.1 GDIP01124954 JAL78760.1 GDIQ01084889 JAN09848.1 ABJB011053834 DS811420 EEC11164.1 GDIQ01229183 JAK22542.1 GDIP01168079 GDIP01165232 LRGB01003155 JAJ55323.1 KZS03945.1 GDIQ01239279 JAK12446.1 GDIP01064752 JAM38963.1 GEFM01005463 JAP70333.1 GDIP01225169 JAI98232.1 GDIP01232074 JAI91327.1 GDIP01078738 JAM24977.1 MWRG01000453 PRD36095.1 GDIQ01035910 JAN58827.1 GDIP01084935 JAM18780.1 GDIP01086542 JAM17173.1 GDIP01031495 JAM72220.1 GDIP01097943 JAM05772.1 GFJQ02005537 JAW01433.1 DS235072 EEB11358.1 GDIQ01172248 JAK79477.1 KZ288236 PBC31381.1 KK575219 KFW09965.1 GFPF01009496 MAA20642.1 GDIQ01009957 JAN84780.1 GDIP01195128 JAJ28274.1 GDIP01094384 JAM09331.1 GDIP01233562 JAI89839.1 GFWV01002350 MAA27080.1 GFJQ02004410 JAW02560.1 GDHC01001662 JAQ16967.1 KQ971311 KYB29466.1 GFPF01005955 MAA17101.1 QOIP01000011 RLU16747.1 KK453894 KFQ75217.1 KK634098 KFV57410.1 KQ435727 KOX77948.1 KL420603 KFW88473.1 GL445145 EFN90111.1 GBBM01000573 JAC34845.1 KQ414657 KOC65701.1 GGLE01000994 MBY05120.1 KK852727 KDR17628.1 KQ434870 KZC09583.1 PYGN01001252 PSN36278.1 KK844969 KFP86750.1 LSYS01008070 OPJ69880.1 IACK01081567 LAA80435.1 KL218157 KFP02852.1 GL440775 EFN65441.1 KL525914 KFO91172.1 KK529928 KFP27703.1 KL224637 KFW62424.1 KL310194 KFP52940.1 KL447872 KFO78046.1 ADTU01001694 ADTU01001695 KB744656 EOA94666.1 GFWV01022972 MAA47699.1 KK734538 KFR09507.1 AERX01059979 KK401037 KFV59999.1 AL844186 KL215167 KFV63658.1 KK718726 KFO58256.1 KL409720 KFQ91416.1 KK502227 KFP21745.1 KL873654 KGL99641.1 KL226349 KFM12525.1 GBYB01004546 JAG74313.1 OPJ69881.1 KL206966 KFV87619.1 KL669044 KFW75387.1 AWGT02002370 OXB70173.1 KL892829 KGL80197.1 QUSF01000067 RLV96807.1 MUZQ01000093 OWK58673.1 QRBI01000238 RMB91285.1 ADON01133313 ADON01133314
RSAL01000008 RVE54040.1 KQ459600 KPI94123.1 AGBW02012844 OWR44349.1 GDIQ01039155 JAN55582.1 GDIQ01039156 JAN55581.1 KK121162 KFM79884.1 KK852482 KDR22930.1 NNAY01000500 OXU27976.1 NEVH01011197 PNF31816.1 GDIP01143631 JAJ79771.1 GL732523 EFX90178.1 IAAA01016543 LAA04952.1 GDIP01186028 JAJ37374.1 GDIP01158433 JAJ64969.1 GDIQ01171207 JAK80518.1 GDIP01142541 JAL61173.1 GDIQ01229184 JAK22541.1 GDIP01124954 JAL78760.1 GDIQ01084889 JAN09848.1 ABJB011053834 DS811420 EEC11164.1 GDIQ01229183 JAK22542.1 GDIP01168079 GDIP01165232 LRGB01003155 JAJ55323.1 KZS03945.1 GDIQ01239279 JAK12446.1 GDIP01064752 JAM38963.1 GEFM01005463 JAP70333.1 GDIP01225169 JAI98232.1 GDIP01232074 JAI91327.1 GDIP01078738 JAM24977.1 MWRG01000453 PRD36095.1 GDIQ01035910 JAN58827.1 GDIP01084935 JAM18780.1 GDIP01086542 JAM17173.1 GDIP01031495 JAM72220.1 GDIP01097943 JAM05772.1 GFJQ02005537 JAW01433.1 DS235072 EEB11358.1 GDIQ01172248 JAK79477.1 KZ288236 PBC31381.1 KK575219 KFW09965.1 GFPF01009496 MAA20642.1 GDIQ01009957 JAN84780.1 GDIP01195128 JAJ28274.1 GDIP01094384 JAM09331.1 GDIP01233562 JAI89839.1 GFWV01002350 MAA27080.1 GFJQ02004410 JAW02560.1 GDHC01001662 JAQ16967.1 KQ971311 KYB29466.1 GFPF01005955 MAA17101.1 QOIP01000011 RLU16747.1 KK453894 KFQ75217.1 KK634098 KFV57410.1 KQ435727 KOX77948.1 KL420603 KFW88473.1 GL445145 EFN90111.1 GBBM01000573 JAC34845.1 KQ414657 KOC65701.1 GGLE01000994 MBY05120.1 KK852727 KDR17628.1 KQ434870 KZC09583.1 PYGN01001252 PSN36278.1 KK844969 KFP86750.1 LSYS01008070 OPJ69880.1 IACK01081567 LAA80435.1 KL218157 KFP02852.1 GL440775 EFN65441.1 KL525914 KFO91172.1 KK529928 KFP27703.1 KL224637 KFW62424.1 KL310194 KFP52940.1 KL447872 KFO78046.1 ADTU01001694 ADTU01001695 KB744656 EOA94666.1 GFWV01022972 MAA47699.1 KK734538 KFR09507.1 AERX01059979 KK401037 KFV59999.1 AL844186 KL215167 KFV63658.1 KK718726 KFO58256.1 KL409720 KFQ91416.1 KK502227 KFP21745.1 KL873654 KGL99641.1 KL226349 KFM12525.1 GBYB01004546 JAG74313.1 OPJ69881.1 KL206966 KFV87619.1 KL669044 KFW75387.1 AWGT02002370 OXB70173.1 KL892829 KGL80197.1 QUSF01000067 RLV96807.1 MUZQ01000093 OWK58673.1 QRBI01000238 RMB91285.1 ADON01133313 ADON01133314
Proteomes
UP000005204
UP000218220
UP000283053
UP000053268
UP000007151
UP000054359
+ More
UP000027135 UP000215335 UP000235965 UP000000305 UP000001555 UP000076858 UP000009046 UP000242457 UP000007266 UP000279307 UP000053105 UP000008237 UP000053825 UP000076502 UP000245037 UP000190648 UP000054308 UP000000311 UP000054081 UP000053760 UP000005205 UP000053605 UP000005207 UP000000437 UP000053875 UP000052976 UP000053283 UP000053119 UP000053858 UP000053286 UP000261460 UP000053584 UP000053258 UP000005203 UP000198419 UP000053641 UP000264840 UP000276834 UP000197619 UP000269221 UP000016666
UP000027135 UP000215335 UP000235965 UP000000305 UP000001555 UP000076858 UP000009046 UP000242457 UP000007266 UP000279307 UP000053105 UP000008237 UP000053825 UP000076502 UP000245037 UP000190648 UP000054308 UP000000311 UP000054081 UP000053760 UP000005205 UP000053605 UP000005207 UP000000437 UP000053875 UP000052976 UP000053283 UP000053119 UP000053858 UP000053286 UP000261460 UP000053584 UP000053258 UP000005203 UP000198419 UP000053641 UP000264840 UP000276834 UP000197619 UP000269221 UP000016666
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JNB9
A0A2H1VW42
A0A2A4KAF8
A0A3S2M9H5
A0A194PMA9
A0A212ESA4
+ More
A0A0P6G371 A0A0N8ECW4 A0A087UR95 A0A067RQM1 A0A232FAV7 A0A2J7QTA8 A0A0P5EG14 E9FRK1 A0A2L2Y9V1 A0A0P5BDK1 A0A0N8ADD8 A0A0P5MZV8 A0A0P5SJV5 A0A0P5HAY8 A0A0P5TRG1 A0A0P6D054 B7PX42 A0A0P5HDB5 A0A0P5CQS7 A0A0P5GN72 A0A0P5Y0G1 A0A131XSP8 A0A0N7ZPJ6 A0A0P4YCL5 A0A0P5XXE8 A0A2P6LBY2 A0A0P6GBD1 A0A0N8CZ14 A0A0P5WGT4 A0A0N8DI44 A0A0N8CUD8 A0A1Z5L217 E0VDA2 A0A0P5LKM9 A0A2A3EK20 A0A093J4S8 A0A224Z3J6 A0A0P6I601 A0A0P5AR78 A0A0P5VYP3 A0A0P4Y249 A0A293LJU0 A0A1Z5L5L5 A0A146MDQ3 A0A139WNM3 A0A224YR83 A0A3L8D8C0 A0A091THR5 A0A093FL99 A0A0N0BIS0 A0A093QHP7 E2B2H0 A0A023GP06 A0A0L7R4L4 A0A2R5L6N5 A0A067RCK0 A0A154PCN1 A0A2P8XW97 A0A091P5C4 A0A1V4JCQ2 A0A2D4I895 A0A091I3T2 E2AM99 A0A091H8L9 A0A091JUT6 A0A093QUK5 A0A091LCQ0 A0A091G9L6 A0A158NQ99 R0JCF4 A0A293N5M7 A0A091W1H8 I3JBC0 A0A093G052 A0A2R8Q8P2 A0A093G3F3 A0A091EMG5 A0A091UL68 A0A091JPR1 A0A0A0B0G1 A0A087RGC1 A0A3B4GPL8 A0A0C9RC82 A0A1V4JCG5 A0A093KBC1 A0A093PTF6 A0A088AAG3 A0A226NQN8 A0A099ZEK4 A0A3Q2V816 A0A3L8S3J3 A0A218UY33 A0A3M0IRN7 U3IUI0
A0A0P6G371 A0A0N8ECW4 A0A087UR95 A0A067RQM1 A0A232FAV7 A0A2J7QTA8 A0A0P5EG14 E9FRK1 A0A2L2Y9V1 A0A0P5BDK1 A0A0N8ADD8 A0A0P5MZV8 A0A0P5SJV5 A0A0P5HAY8 A0A0P5TRG1 A0A0P6D054 B7PX42 A0A0P5HDB5 A0A0P5CQS7 A0A0P5GN72 A0A0P5Y0G1 A0A131XSP8 A0A0N7ZPJ6 A0A0P4YCL5 A0A0P5XXE8 A0A2P6LBY2 A0A0P6GBD1 A0A0N8CZ14 A0A0P5WGT4 A0A0N8DI44 A0A0N8CUD8 A0A1Z5L217 E0VDA2 A0A0P5LKM9 A0A2A3EK20 A0A093J4S8 A0A224Z3J6 A0A0P6I601 A0A0P5AR78 A0A0P5VYP3 A0A0P4Y249 A0A293LJU0 A0A1Z5L5L5 A0A146MDQ3 A0A139WNM3 A0A224YR83 A0A3L8D8C0 A0A091THR5 A0A093FL99 A0A0N0BIS0 A0A093QHP7 E2B2H0 A0A023GP06 A0A0L7R4L4 A0A2R5L6N5 A0A067RCK0 A0A154PCN1 A0A2P8XW97 A0A091P5C4 A0A1V4JCQ2 A0A2D4I895 A0A091I3T2 E2AM99 A0A091H8L9 A0A091JUT6 A0A093QUK5 A0A091LCQ0 A0A091G9L6 A0A158NQ99 R0JCF4 A0A293N5M7 A0A091W1H8 I3JBC0 A0A093G052 A0A2R8Q8P2 A0A093G3F3 A0A091EMG5 A0A091UL68 A0A091JPR1 A0A0A0B0G1 A0A087RGC1 A0A3B4GPL8 A0A0C9RC82 A0A1V4JCG5 A0A093KBC1 A0A093PTF6 A0A088AAG3 A0A226NQN8 A0A099ZEK4 A0A3Q2V816 A0A3L8S3J3 A0A218UY33 A0A3M0IRN7 U3IUI0
PDB
5YJB
E-value=3.12166e-140,
Score=1278
Ontologies
GO
GO:0016491
GO:0003677
GO:0008168
GO:0050660
GO:0034720
GO:0006355
GO:0005634
GO:0008134
GO:0000122
GO:0003682
GO:0045944
GO:0034648
GO:0000790
GO:1903706
GO:0007399
GO:1900052
GO:0000784
GO:1903827
GO:0071480
GO:0019899
GO:0043518
GO:1902166
GO:0050681
GO:0043426
GO:0032454
GO:0010569
GO:0002039
GO:0034644
GO:0002052
GO:0005654
GO:0042162
GO:0030374
GO:0032091
GO:0061752
GO:0051091
GO:0033184
GO:1990391
GO:0043433
GO:0005515
GO:0051276
GO:0016620
GO:0016787
GO:0003676
GO:0015930
GO:0055114
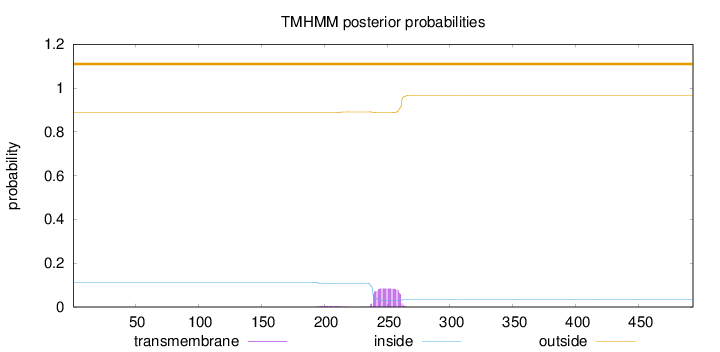
Topology
Subcellular location
Nucleus
Length:
493
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.93415
Exp number, first 60 AAs:
0
Total prob of N-in:
0.11275
outside
1 - 493
Population Genetic Test Statistics
Pi
170.162057
Theta
147.101099
Tajima's D
1.311433
CLR
106.619063
CSRT
0.738763061846908
Interpretation
Uncertain
