Gene
KWMTBOMO13688
Annotation
endonuclease_and_reverse_transcriptase-like_protein_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 1.986
Sequence
CDS
ATGACCATTAACGCTGGCGTTCCACAAGGTTCGGTGCTCTCCCCCACGCTTTTCATCCTGTATATCAATGACATGCTCTCTATTGATGGCATGCATTGCTATGCGGATGACAGCACGGGGGATGCGCGATATATCGGCCATCAGAGTCTTTCTCGGAGCGCGGTGCAAGAGAGACGATCAAAACTTGTGTCTGAAGTGGAGAACTCTCTGGGGCGAGTCTCTGAATGGGGTAAATTGAACTTGGTTCAATTCAACCCTATAAAGACACAAGTTTGCGCGTTCACTGCGAAGAAGGACCCCTTTGTCATGGCGCCGCAATTCCAAGGAGTATCCCTACAACCTTCCGAGAGTATCGGGATACTTGGGGTCGACATTTCGAGCGATGTCCAGTTTCGGAGTCATTTGGAAGGCAAAGCCAAGTTGGCGTCCAAAATGCTGGGAGTCCTCAACAGAGCGAAGCGGTACTTCACGCCTGGACAAAGACTTTTGCTTTATAAAGCACAAGTCCGGCCTCGCGTGGAGTACTGCTCCCATCTCTGGGCCGGGGCTCCCAAATACCAGCTTCTTCCATTTGACTCCATACAGAGGAGGGCCGTTCGGATTGTCGATAATCCCATTCTCTCGGATCGTTTGGAGCCTCTCGGTCTGCGGAGGGACTTCGGTTCCCTCTGTATTTTGTACCGTATGTTCCATGGGGAGTGCTCTGAGGAATTGTTCGAGATGATACCGGCATCTCGTTTTTACCATCGCACTGCCCGCCACCGGAATAGAGTTCATCCATACTACCTGGAGCCACTGCGGTCATCCACACACCTGGTCCCTCGTTTGTTGGGGGTGGCCGGCGCGTTAAGCCGGCTTCTTCCCAACATCGGGGGGCCTGACCAGGTGACGCGCCGTCTCTATACGGGGGTGGTGCGGTCAATGGCCTTATACGGGGCACCCGTGTGGGGCCAGTCCCTGGCCGTGGGGGTAGCGAAGTTGCTGCAACGGCCGCAACGCACCATCGCGGTCAGGGTCATCCGTGGTTATCGCACCATCTCTTTTGAGGCGGCGTGTGTACTGGCTGGGACGCCGCCTTGGGTCCTGGAGGCGGAGGCGCTCGCCGCTGACTATCAGTGGCGGGCTAACCTTCGTGCCCGGGGCGTGGCGCATCCCAGCCCCAGTGTGGTCAGAGCGCGGAGGGCCCAATCTCGGCGGTCCGTGCTGGAGTCATGGTCCAGGCGGCTGGCCCATCCTTCGGCTGGTCGTAGGACCGTCGAGGCGATTCGCCCGATTCTTGTGGATTGGGTGAATCGTGACAGAGGACGCCTCACTTTCCGGCTCACGCAGGTGCTCACTGGGCATGGTTGCTTCGGTGAGTTCCTGCACCGGATCGGAGCCGAGCCGACGGCAGAGTGTTACCATTGTGGTTGCGACTTGGACACGGCGGAGCACACGCTCATTGCCTGCCCCGCATGGGAGGGGTGGCGCCGTGTCCTTGCCGCAAAAATAGGAAACGACTTGTCGTTGCCGAGTGTTGTGGCATCGATGCTCGGCAACGACGAGTCGTGGAAGGCGATGCTCGACTTCTGCGAGTGCACCATCTCGCAGAAGGAGGCGGCGGGGCGCGTGAGAGACGCACAAGTCCGCCGCCGTCGAGCGGGGGCCAGGGAGGCGGATCTCGCCCTAGCCCTGGCCCTCTAA
Protein
MTINAGVPQGSVLSPTLFILYINDMLSIDGMHCYADDSTGDARYIGHQSLSRSAVQERRSKLVSEVENSLGRVSEWGKLNLVQFNPIKTQVCAFTAKKDPFVMAPQFQGVSLQPSESIGILGVDISSDVQFRSHLEGKAKLASKMLGVLNRAKRYFTPGQRLLLYKAQVRPRVEYCSHLWAGAPKYQLLPFDSIQRRAVRIVDNPILSDRLEPLGLRRDFGSLCILYRMFHGECSEELFEMIPASRFYHRTARHRNRVHPYYLEPLRSSTHLVPRLLGVAGALSRLLPNIGGPDQVTRRLYTGVVRSMALYGAPVWGQSLAVGVAKLLQRPQRTIAVRVIRGYRTISFEAACVLAGTPPWVLEAEALAADYQWRANLRARGVAHPSPSVVRARRAQSRRSVLESWSRRLAHPSAGRRTVEAIRPILVDWVNRDRGRLTFRLTQVLTGHGCFGEFLHRIGAEPTAECYHCGCDLDTAEHTLIACPAWEGWRRVLAAKIGNDLSLPSVVASMLGNDESWKAMLDFCECTISQKEAAGRVRDAQVRRRRAGAREADLALALAL
Summary
Uniprot
ProteinModelPortal
Ontologies
GO
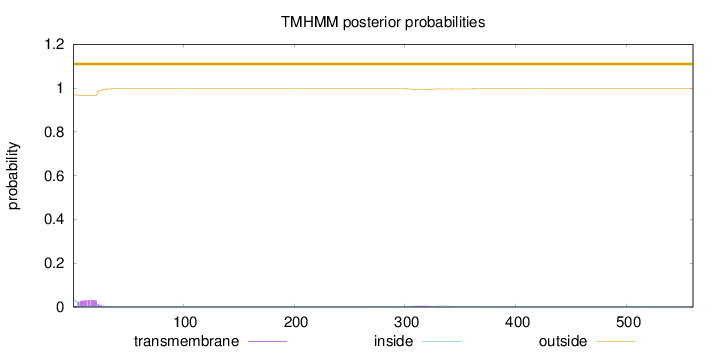
Topology
Length:
560
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.730159999999998
Exp number, first 60 AAs:
0.59979
Total prob of N-in:
0.03217
outside
1 - 560
Population Genetic Test Statistics
Pi
197.340618
Theta
170.58282
Tajima's D
1.174829
CLR
0.476733
CSRT
0.708864556772161
Interpretation
Uncertain
