Gene
KWMTBOMO13686
Pre Gene Modal
BGIBMGA011023
Annotation
PREDICTED:_lysine-specific_histone_demethylase_1A_[Bombyx_mori]
Full name
Probable lysine-specific histone demethylase 1
+ More
Lysine-specific histone demethylase
Lysine-specific histone demethylase
Location in the cell
PlasmaMembrane Reliability : 1.591
Sequence
CDS
ATGTCGTTAAAGTGTGATGATAACGCTAAAGAGTACAAAATAACGTACTTTCGTAACGGGTATTCCTGCGTGCCCGTCGCTCTGTCAGAAGACTTGGACATCAGGCTCGGGACCTCGGTAACTGATATCAACTACGAAGGCCCAGGAGTAACAGTTAAAGCTAATAGTTCTCGCAATCCGAACCAGCCACAGGTCTTTAAAGGAGATGTAGTCTTATGTACATTGCCTTTGGGAGTGTTAAAGGTTGCAGTGGCAAGCAATGGACAGAATCAACAGAACTTCACCACTTTTAATCCCCCCCTGCCCGAGTGGAAGGTGGCAGCTATCAAGCGTCTCGGTTATGGAAATTTGAACAAAGTGGTTTTGTGCTTCGAACGAACCTTCTGGGACCCGAGTGCCAATCTATTTGGGCACGTTGGTACGACCACCGCAAGCAGAGGAGAACTTTTCCTTTTCTGGAACCTCTATAGTGCGCCGGTACTCCTAGCTTTGGTAGCAGGAGAGGCAGCGGCTGTAATGGAAAACGTCACCGATGACGTCATCGTCGGTAGATGTATCGCCGTACTGAAGAGCATATTTGGTCACGCGGCCGTGCCGCAACCCAAAGAATGCATAGTAACTAGATGGCGTGCCGATCCATTCGCTCGCGGCTCGTACAGTTTTGTAGCGGTAGGGTCGTCTGGTACTGACTACGATCTCCTGGCCGCCCCTATACCTGGAACTGATGGAGAGAATAGACTTTTCTTTGCCGGTGAGCACACGATGCGGAACTATCCAGCAACGGTGCACGGAGCATTCTTATCGGGTTTAAGGGAAGCTGGGAGATTGGCCGACATGTTGCTTCCTCTGCCACCAATCACTAGCTCGAGTGTCGCTGCCGTGACGGCAAACAATTCCGCATGA
Protein
MSLKCDDNAKEYKITYFRNGYSCVPVALSEDLDIRLGTSVTDINYEGPGVTVKANSSRNPNQPQVFKGDVVLCTLPLGVLKVAVASNGQNQQNFTTFNPPLPEWKVAAIKRLGYGNLNKVVLCFERTFWDPSANLFGHVGTTTASRGELFLFWNLYSAPVLLALVAGEAAAVMENVTDDVIVGRCIAVLKSIFGHAAVPQPKECIVTRWRADPFARGSYSFVAVGSSGTDYDLLAAPIPGTDGENRLFFAGEHTMRNYPATVHGAFLSGLREAGRLADMLLPLPPITSSSVAAVTANNSA
Summary
Description
Probable histone demethylase that specifically demethylates 'Lys-4' of histone H3, a specific tag for epigenetic transcriptional activation, thereby acting as a corepressor. Acts by oxidizing the substrate by FAD to generate the corresponding imine that is subsequently hydrolyzed. Demethylates both mono- and di-methylated 'Lys-4' of histone H3.
Histone demethylase that demethylates both 'Lys-4' (H3K4me) and 'Lys-9' (H3K9me) of histone H3, thereby acting as a coactivator or a corepressor, depending on the context. Acts by oxidizing the substrate by FAD to generate the corresponding imine that is subsequently hydrolyzed. Acts as a corepressor by mediating demethylation of H3K4me, a specific tag for epigenetic transcriptional activation. Demethylates both mono- (H3K4me1) and di-methylated (H3K4me2) H3K4me.
Histone demethylase that demethylates both 'Lys-4' (H3K4me) and 'Lys-9' (H3K9me) of histone H3, thereby acting as a coactivator or a corepressor, depending on the context. Acts by oxidizing the substrate by FAD to generate the corresponding imine that is subsequently hydrolyzed. Acts as a corepressor by mediating demethylation of H3K4me, a specific tag for epigenetic transcriptional activation. Demethylates both mono- (H3K4me1) and di-methylated (H3K4me2) H3K4me.
Cofactor
FAD
Similarity
Belongs to the flavin monoamine oxidase family.
Feature
chain Lysine-specific histone demethylase
Uniprot
A0A194PL11
A0A2A4K937
A0A212ES90
A0A194RER2
A0A2H1V084
A0A1B6CNV3
+ More
A0A1B6F811 A0A1B6LH74 E0VDA5 A0A2P8XW97 A0A139WNH8 A0A067RCK0 A0A0T6B3R9 A0A1W4WU62 A0A2T7PGM5 A0A2J7QZ86 T1HJ69 A0A0S7LFX9 A0A1Y1LPN4 A0A2I4C8Q9 A0A146PHN5 A0A1A8HYI7 A0A0P4WIF3 A0A2D4FJW5 C3RZ91 G3P5A9 K1PSW2 K9KDA9 A0A1A8CDQ2 A0A091PYN2 A0A026W5K8 V3ZE46 A0A2D4FJX6 A0A3L8D8C0 A0A087VAN2 A0A2H8TSG7 A0A2I0TTJ3 A0A1A8FXG4 A0A210QFJ1 K7JWZ3 K9IT43 A0A0A9XBP8 J9LER4 A0A2K6G369 A0A2I3GMX2 G2HF04 A0A069DWN6 A0A3L7I8K4 A0A147A4N6 A0A087UR95 A0A0P5C548 A0A0P5YFY9 A0A3P8PZC4 A0A3P9B3Z1 A0A1E1XDE1 A0A0L8IEY1 A0A0P5C2W2 A0A0P5XQ71 A0A023F516 A0A2L2Y9V1 A0A3P8TNG1 A0A3Q1BH70 A0A3B4GQ09 A0A0P5UZB8 M4A829 A0A3P9NB14 A0A3B3XUA6 A0A0P5YP84 A0A0P4W046 V9KEM0 A0A3B3SYF8 A0A3P9LB88 G3P5B5 A0A146WGX6 A0A3Q0SCS9 A0A3B4YR59 A0A1A6GE51 A0A3Q2FX39 A0A3Q2XZ25 S9Y4V3 A0A2R5L6N5 A0A293N5M7 A0A3Q2QKI9 A0A131YYD9 A0A091G9L6 H2MB66 I3JBC0 A0A0L7R4F2 A0A093PTF6 A0A3P9LBI8 A0A3Q3LKE1 A0A091JUT6 A0A093QUK5 A0A3B3UW04 A0A3Q2E7G6 A0A146WJI3 A0A091LCQ0 A0A3Q2V816 A0A3P9NAZ4
A0A1B6F811 A0A1B6LH74 E0VDA5 A0A2P8XW97 A0A139WNH8 A0A067RCK0 A0A0T6B3R9 A0A1W4WU62 A0A2T7PGM5 A0A2J7QZ86 T1HJ69 A0A0S7LFX9 A0A1Y1LPN4 A0A2I4C8Q9 A0A146PHN5 A0A1A8HYI7 A0A0P4WIF3 A0A2D4FJW5 C3RZ91 G3P5A9 K1PSW2 K9KDA9 A0A1A8CDQ2 A0A091PYN2 A0A026W5K8 V3ZE46 A0A2D4FJX6 A0A3L8D8C0 A0A087VAN2 A0A2H8TSG7 A0A2I0TTJ3 A0A1A8FXG4 A0A210QFJ1 K7JWZ3 K9IT43 A0A0A9XBP8 J9LER4 A0A2K6G369 A0A2I3GMX2 G2HF04 A0A069DWN6 A0A3L7I8K4 A0A147A4N6 A0A087UR95 A0A0P5C548 A0A0P5YFY9 A0A3P8PZC4 A0A3P9B3Z1 A0A1E1XDE1 A0A0L8IEY1 A0A0P5C2W2 A0A0P5XQ71 A0A023F516 A0A2L2Y9V1 A0A3P8TNG1 A0A3Q1BH70 A0A3B4GQ09 A0A0P5UZB8 M4A829 A0A3P9NB14 A0A3B3XUA6 A0A0P5YP84 A0A0P4W046 V9KEM0 A0A3B3SYF8 A0A3P9LB88 G3P5B5 A0A146WGX6 A0A3Q0SCS9 A0A3B4YR59 A0A1A6GE51 A0A3Q2FX39 A0A3Q2XZ25 S9Y4V3 A0A2R5L6N5 A0A293N5M7 A0A3Q2QKI9 A0A131YYD9 A0A091G9L6 H2MB66 I3JBC0 A0A0L7R4F2 A0A093PTF6 A0A3P9LBI8 A0A3Q3LKE1 A0A091JUT6 A0A093QUK5 A0A3B3UW04 A0A3Q2E7G6 A0A146WJI3 A0A091LCQ0 A0A3Q2V816 A0A3P9NAZ4
EC Number
1.-.-.-
Pubmed
EMBL
KQ459600
KPI94121.1
NWSH01000020
PCG80717.1
AGBW02012844
OWR44347.1
+ More
KQ460473 KPJ14421.1 ODYU01000058 SOQ34186.1 GEDC01022148 JAS15150.1 GECZ01023736 JAS46033.1 GEBQ01016932 JAT23045.1 DS235072 EEB11361.1 PYGN01001252 PSN36278.1 KQ971311 KYB29464.1 KK852727 KDR17628.1 LJIG01016034 KRT81813.1 PZQS01000004 PVD32566.1 NEVH01009072 PNF33900.1 ACPB03003082 GBYX01171309 GBYX01171306 GBYX01171303 JAO88370.1 GEZM01050266 GEZM01050265 JAV75644.1 GCES01142891 JAQ43431.1 HAED01003630 SBQ89618.1 GDRN01087837 JAI60978.1 IACJ01076167 LAA47763.1 EU661933 ACH71255.1 JH818227 EKC24738.1 JL623189 AEP99694.1 HADZ01013858 SBP77799.1 KK680533 KFQ12431.1 KK107403 EZA51288.1 KB202619 ESO89368.1 IACJ01076170 LAA47773.1 QOIP01000011 RLU16747.1 KL486279 KFO09674.1 GFXV01004343 MBW16148.1 KZ507299 PKU37124.1 HAEB01016939 SBQ63466.1 NEDP02003873 OWF47510.1 AAZX01007402 AAZX01008991 AAZX01010111 AAZX01020796 GABZ01001325 JAA52200.1 GBHO01027367 GBHO01027366 GBRD01005696 GBRD01005695 JAG16237.1 JAG16238.1 JAG60125.1 ABLF02038378 ADFV01066899 ADFV01066900 AK305318 BAK62312.1 GBGD01000772 JAC88117.1 RAZU01000087 RLQ74525.1 GCES01013171 JAR73152.1 KK121162 KFM79884.1 GDIP01180078 JAJ43324.1 GDIP01058244 JAM45471.1 GFAC01001923 JAT97265.1 KQ415861 KOG00015.1 GDIP01191877 JAJ31525.1 GDIP01081243 JAM22472.1 GBBI01002569 JAC16143.1 IAAA01016543 LAA04952.1 GDIP01106692 JAL97022.1 GDIP01056385 JAM47330.1 GDKW01001469 JAI55126.1 JW863742 AFO96259.1 GCES01057083 JAR29240.1 LZPO01099511 OBS63602.1 KB016923 EPY82441.1 GGLE01000994 MBY05120.1 GFWV01022972 MAA47699.1 GEDV01005037 JAP83520.1 KL447872 KFO78046.1 AERX01059979 KQ414657 KOC65704.1 KL669044 KFW75387.1 KK529928 KFP27703.1 KL224637 KFW62424.1 GCES01057081 JAR29242.1 KL310194 KFP52940.1
KQ460473 KPJ14421.1 ODYU01000058 SOQ34186.1 GEDC01022148 JAS15150.1 GECZ01023736 JAS46033.1 GEBQ01016932 JAT23045.1 DS235072 EEB11361.1 PYGN01001252 PSN36278.1 KQ971311 KYB29464.1 KK852727 KDR17628.1 LJIG01016034 KRT81813.1 PZQS01000004 PVD32566.1 NEVH01009072 PNF33900.1 ACPB03003082 GBYX01171309 GBYX01171306 GBYX01171303 JAO88370.1 GEZM01050266 GEZM01050265 JAV75644.1 GCES01142891 JAQ43431.1 HAED01003630 SBQ89618.1 GDRN01087837 JAI60978.1 IACJ01076167 LAA47763.1 EU661933 ACH71255.1 JH818227 EKC24738.1 JL623189 AEP99694.1 HADZ01013858 SBP77799.1 KK680533 KFQ12431.1 KK107403 EZA51288.1 KB202619 ESO89368.1 IACJ01076170 LAA47773.1 QOIP01000011 RLU16747.1 KL486279 KFO09674.1 GFXV01004343 MBW16148.1 KZ507299 PKU37124.1 HAEB01016939 SBQ63466.1 NEDP02003873 OWF47510.1 AAZX01007402 AAZX01008991 AAZX01010111 AAZX01020796 GABZ01001325 JAA52200.1 GBHO01027367 GBHO01027366 GBRD01005696 GBRD01005695 JAG16237.1 JAG16238.1 JAG60125.1 ABLF02038378 ADFV01066899 ADFV01066900 AK305318 BAK62312.1 GBGD01000772 JAC88117.1 RAZU01000087 RLQ74525.1 GCES01013171 JAR73152.1 KK121162 KFM79884.1 GDIP01180078 JAJ43324.1 GDIP01058244 JAM45471.1 GFAC01001923 JAT97265.1 KQ415861 KOG00015.1 GDIP01191877 JAJ31525.1 GDIP01081243 JAM22472.1 GBBI01002569 JAC16143.1 IAAA01016543 LAA04952.1 GDIP01106692 JAL97022.1 GDIP01056385 JAM47330.1 GDKW01001469 JAI55126.1 JW863742 AFO96259.1 GCES01057083 JAR29240.1 LZPO01099511 OBS63602.1 KB016923 EPY82441.1 GGLE01000994 MBY05120.1 GFWV01022972 MAA47699.1 GEDV01005037 JAP83520.1 KL447872 KFO78046.1 AERX01059979 KQ414657 KOC65704.1 KL669044 KFW75387.1 KK529928 KFP27703.1 KL224637 KFW62424.1 GCES01057081 JAR29242.1 KL310194 KFP52940.1
Proteomes
UP000053268
UP000218220
UP000007151
UP000053240
UP000009046
UP000245037
+ More
UP000007266 UP000027135 UP000192223 UP000245119 UP000235965 UP000015103 UP000192220 UP000007635 UP000005408 UP000053097 UP000030746 UP000279307 UP000242188 UP000002358 UP000007819 UP000233160 UP000001073 UP000273346 UP000054359 UP000265100 UP000265160 UP000053454 UP000265080 UP000257160 UP000261460 UP000002852 UP000242638 UP000261480 UP000261540 UP000265180 UP000261340 UP000261360 UP000092124 UP000265020 UP000264820 UP000265000 UP000053760 UP000001038 UP000005207 UP000053825 UP000053258 UP000261640 UP000054081 UP000261500 UP000264840
UP000007266 UP000027135 UP000192223 UP000245119 UP000235965 UP000015103 UP000192220 UP000007635 UP000005408 UP000053097 UP000030746 UP000279307 UP000242188 UP000002358 UP000007819 UP000233160 UP000001073 UP000273346 UP000054359 UP000265100 UP000265160 UP000053454 UP000265080 UP000257160 UP000261460 UP000002852 UP000242638 UP000261480 UP000261540 UP000265180 UP000261340 UP000261360 UP000092124 UP000265020 UP000264820 UP000265000 UP000053760 UP000001038 UP000005207 UP000053825 UP000053258 UP000261640 UP000054081 UP000261500 UP000264840
Interpro
Gene 3D
ProteinModelPortal
A0A194PL11
A0A2A4K937
A0A212ES90
A0A194RER2
A0A2H1V084
A0A1B6CNV3
+ More
A0A1B6F811 A0A1B6LH74 E0VDA5 A0A2P8XW97 A0A139WNH8 A0A067RCK0 A0A0T6B3R9 A0A1W4WU62 A0A2T7PGM5 A0A2J7QZ86 T1HJ69 A0A0S7LFX9 A0A1Y1LPN4 A0A2I4C8Q9 A0A146PHN5 A0A1A8HYI7 A0A0P4WIF3 A0A2D4FJW5 C3RZ91 G3P5A9 K1PSW2 K9KDA9 A0A1A8CDQ2 A0A091PYN2 A0A026W5K8 V3ZE46 A0A2D4FJX6 A0A3L8D8C0 A0A087VAN2 A0A2H8TSG7 A0A2I0TTJ3 A0A1A8FXG4 A0A210QFJ1 K7JWZ3 K9IT43 A0A0A9XBP8 J9LER4 A0A2K6G369 A0A2I3GMX2 G2HF04 A0A069DWN6 A0A3L7I8K4 A0A147A4N6 A0A087UR95 A0A0P5C548 A0A0P5YFY9 A0A3P8PZC4 A0A3P9B3Z1 A0A1E1XDE1 A0A0L8IEY1 A0A0P5C2W2 A0A0P5XQ71 A0A023F516 A0A2L2Y9V1 A0A3P8TNG1 A0A3Q1BH70 A0A3B4GQ09 A0A0P5UZB8 M4A829 A0A3P9NB14 A0A3B3XUA6 A0A0P5YP84 A0A0P4W046 V9KEM0 A0A3B3SYF8 A0A3P9LB88 G3P5B5 A0A146WGX6 A0A3Q0SCS9 A0A3B4YR59 A0A1A6GE51 A0A3Q2FX39 A0A3Q2XZ25 S9Y4V3 A0A2R5L6N5 A0A293N5M7 A0A3Q2QKI9 A0A131YYD9 A0A091G9L6 H2MB66 I3JBC0 A0A0L7R4F2 A0A093PTF6 A0A3P9LBI8 A0A3Q3LKE1 A0A091JUT6 A0A093QUK5 A0A3B3UW04 A0A3Q2E7G6 A0A146WJI3 A0A091LCQ0 A0A3Q2V816 A0A3P9NAZ4
A0A1B6F811 A0A1B6LH74 E0VDA5 A0A2P8XW97 A0A139WNH8 A0A067RCK0 A0A0T6B3R9 A0A1W4WU62 A0A2T7PGM5 A0A2J7QZ86 T1HJ69 A0A0S7LFX9 A0A1Y1LPN4 A0A2I4C8Q9 A0A146PHN5 A0A1A8HYI7 A0A0P4WIF3 A0A2D4FJW5 C3RZ91 G3P5A9 K1PSW2 K9KDA9 A0A1A8CDQ2 A0A091PYN2 A0A026W5K8 V3ZE46 A0A2D4FJX6 A0A3L8D8C0 A0A087VAN2 A0A2H8TSG7 A0A2I0TTJ3 A0A1A8FXG4 A0A210QFJ1 K7JWZ3 K9IT43 A0A0A9XBP8 J9LER4 A0A2K6G369 A0A2I3GMX2 G2HF04 A0A069DWN6 A0A3L7I8K4 A0A147A4N6 A0A087UR95 A0A0P5C548 A0A0P5YFY9 A0A3P8PZC4 A0A3P9B3Z1 A0A1E1XDE1 A0A0L8IEY1 A0A0P5C2W2 A0A0P5XQ71 A0A023F516 A0A2L2Y9V1 A0A3P8TNG1 A0A3Q1BH70 A0A3B4GQ09 A0A0P5UZB8 M4A829 A0A3P9NB14 A0A3B3XUA6 A0A0P5YP84 A0A0P4W046 V9KEM0 A0A3B3SYF8 A0A3P9LB88 G3P5B5 A0A146WGX6 A0A3Q0SCS9 A0A3B4YR59 A0A1A6GE51 A0A3Q2FX39 A0A3Q2XZ25 S9Y4V3 A0A2R5L6N5 A0A293N5M7 A0A3Q2QKI9 A0A131YYD9 A0A091G9L6 H2MB66 I3JBC0 A0A0L7R4F2 A0A093PTF6 A0A3P9LBI8 A0A3Q3LKE1 A0A091JUT6 A0A093QUK5 A0A3B3UW04 A0A3Q2E7G6 A0A146WJI3 A0A091LCQ0 A0A3Q2V816 A0A3P9NAZ4
PDB
2HKO
E-value=5.82638e-96,
Score=894
Ontologies
GO
Topology
Subcellular location
Nucleus
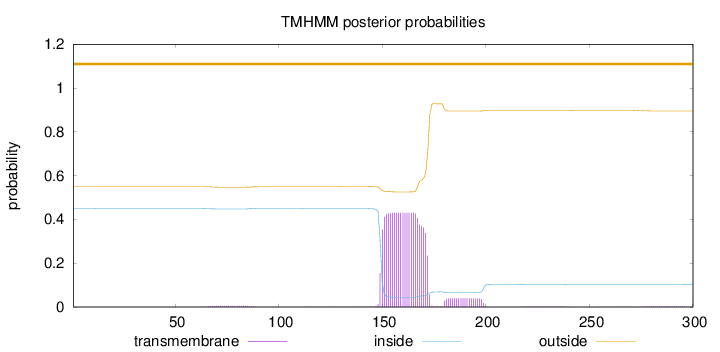
Length:
300
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
10.53759
Exp number, first 60 AAs:
0.00115
Total prob of N-in:
0.44953
outside
1 - 300
Population Genetic Test Statistics
Pi
226.507006
Theta
167.552819
Tajima's D
1.402522
CLR
0.069962
CSRT
0.76341182940853
Interpretation
Uncertain
