Gene
KWMTBOMO13680
Annotation
PREDICTED:_uncharacterized_protein_LOC105385758_[Plutella_xylostella]
Location in the cell
PlasmaMembrane Reliability : 1.362
Sequence
CDS
ATGGTGGCCGAGGAACTTAAAAAAAACATTGATCAAGTTTTGCATGACAATAATGTCACAAGTTGGGAGGATCTTTTATCATTTGCATATAGAATTTTGCATGTAGATAAACAGAGCAATAACTCCCATTCACTTACTAAAAAAATTAAGGATAATTGTAAGAATAATTCCAATAAGACTTTGTTGGACGATGGGGTAAATTATGACAAGCAACAGCGTCCATTTAATAAAGTTGCAAAAATTGAGGGGAAAGTAAGTGATGGAGATTTAAAGGGTGCCTCACAACTATTATTTTCATCAGATTCTCTAGCTCCGAATAATGCCGACACTCTTCTGGCGTTGAAGTCGAAACATCCTGCACCTTTAACACCTGTTAGTGTGCCGCCACCACAGTCTTCCACAACTTTGCCTCGTGTGTCTCTTGATGACGTTCTCACCACAATTACATCTTTTAAAAACGGTTCAGCAGCCGGACTGGATGGTCTATCCCCGCAGCATTTCAAGGATTTAATCAATGTTTCTGCTGGTGATGTCGGGCGGTCTTTACTTAAAAGTATAGTCTCGCTTGTTAATTTAATGCTGGCGGGAAGTGTACCTGAAAGTATCGCTGATATTTTGTATGGAGCGAATTTGTGTGCTTTGATGAAGAGAGATGGCGGTATTAGGCCTATCGCTGTTGGCGGTACACTGCGCCGCATTGTATCCAGGCTTTGCTGCAAAGAGGTTCTGCCGATATTGAGTGGGATGTTCCGGCCGATACAGTTGGGCGTCGGTACAAAGGGCGGCTGTGAAGCTGCGGTACATGCGGCTCGCACCTTTCTTAAGGGGGGTGGTGCCGAAGTATTGGTGAAGGTGGATTTAAAAAATGCATTTAATTGTATCGATAGGAATGCTTTTCTCAAAGAAGTGAAGGAACACATACCATCTATCTACCCTTACCTTTATCAATGCTATGGTAATCCATCTAAGCTGGTTTATAAGGAGAACCTTGTGGAATCAGCGACGGGTTGTCAACAAGGTGATCCACTCGGTCCGGCGATTTTTAGTTTGGGGATACATCCAGTTTTGAGGGAGCTGAAGTCCAGCCTAAATTTATGGTATCTGGATGACGGAACATTGGGAGGGGACATTTCCACGGTCTTAAGGGATTTAGAATATCTAAAATCTGCTTTGGCCAGTTTGGGTTTAGAGCTCAATTTTGCTAAGTGTGAGCTTTTTATCAATGCCTCTTGTTGTTCCGTACCAGTTTCTGACGTTTTTGCAAAGTTTAATACAGCAGCGCCCAACATAAAAATTGTTGATGAGAATTCTCTGATGTTGCTGGGTGCTCCCATTTTGGATGGATCGATCGACGCGTATATTGATGAGCAGATTGATAAATTTTCTCAGTGTTCCAACAATTTATTTGAAATTAACTCTCATATGGCTTTGTTCATCATAAGACACTGTCTATTCGTACCTAAATTTACATATGTGTTAAGATGTTCTTTCTTATGGAAACATAATAATAAACTTAATTCTTTGGATGAAATGATAAAAGGTGTTCTGTGCAAAATAATTAATTGTAATTTGGGTGACCTCTCTTGGGAGCAATGTGTCCTGCCGGTACGTTTTGGTGGTATCGGCGTACGTAAAGCATCAGCTGTTGCATTGCCGGCCTTTCTTGCTTCTGCACACGCCTCCTGTCTCATAGTCGGTAATGTTCTCAACCCTTGTATGGGTTCCGTAGAGATTGTGGGTGTGTCGGAGGCTGGATCGGCCTGGCAGTCTTTGTGTCAGGCGGATTTTCCTACTTTACCGCATTTGCAGCGTCTTTGGGACACTCCGCTTTGTCAACGGATACAGCAACGGTTACTACAGTCATATGACAACAGCGTGGACATTGCTCGTTTTCGCGCCGTTACCGAAAAGGAATCAAGTTACTGGCTTCAAGCCTATCCATCGTCCAATACAGGCACGTTGTTGGACAACAACGCTCTCTCGCTAGCTATTGGCTTACGGTTGGGGGCCCATATTAACGTACCACATTTCTGTGTCTGTGGCGAGTACGTCAATGAGCTTGGACATCACGGACTCTCGTGCCAGAAAAGCGCTGGGCGCATTCCAAGACACGCGTCTTTAAATGATATGATAAGAAGGGCGCTCACTACAATCAATGTTCCGGCTGTCTTGGAACCCATCGGAATTTTGAGGTCAGATGGGAAAAGGCCCGATGGAGTCACGCTGATTCCATGGTCAGTTGGTAGGTGCCTGGTATGGGATGCTACTTGTGTGGATACTGTGGCTCCCTGTCATTTAGATGGTACTTCGTCCCGCCCCGGTTCAGCTGCAGCTTCAGCAGAAATTACAAAAAAGCGGAAGTATTCGGGTTTGGGGTCTTCTTATTCTTTTTTGCCTTTCGCGGTTGAGACGATGGGCCCTTGGGGCCCTGAAGCAAAGAAATTTTTTACAGAAATCTCCAAACGCTTGAAGGAGGTTACACATGATGCTAAGGCTGGCTGGTACTTTGCACAGAGAGTGAGTCTGGCTGTCCAACGAGGCAACGTAGCCAGTATCTTGGGGACTGTGCCTCCTTCAAGCGCGTTGGAAGATCTGTTCTACTGTGTGGGTTGA
Protein
MVAEELKKNIDQVLHDNNVTSWEDLLSFAYRILHVDKQSNNSHSLTKKIKDNCKNNSNKTLLDDGVNYDKQQRPFNKVAKIEGKVSDGDLKGASQLLFSSDSLAPNNADTLLALKSKHPAPLTPVSVPPPQSSTTLPRVSLDDVLTTITSFKNGSAAGLDGLSPQHFKDLINVSAGDVGRSLLKSIVSLVNLMLAGSVPESIADILYGANLCALMKRDGGIRPIAVGGTLRRIVSRLCCKEVLPILSGMFRPIQLGVGTKGGCEAAVHAARTFLKGGGAEVLVKVDLKNAFNCIDRNAFLKEVKEHIPSIYPYLYQCYGNPSKLVYKENLVESATGCQQGDPLGPAIFSLGIHPVLRELKSSLNLWYLDDGTLGGDISTVLRDLEYLKSALASLGLELNFAKCELFINASCCSVPVSDVFAKFNTAAPNIKIVDENSLMLLGAPILDGSIDAYIDEQIDKFSQCSNNLFEINSHMALFIIRHCLFVPKFTYVLRCSFLWKHNNKLNSLDEMIKGVLCKIINCNLGDLSWEQCVLPVRFGGIGVRKASAVALPAFLASAHASCLIVGNVLNPCMGSVEIVGVSEAGSAWQSLCQADFPTLPHLQRLWDTPLCQRIQQRLLQSYDNSVDIARFRAVTEKESSYWLQAYPSSNTGTLLDNNALSLAIGLRLGAHINVPHFCVCGEYVNELGHHGLSCQKSAGRIPRHASLNDMIRRALTTINVPAVLEPIGILRSDGKRPDGVTLIPWSVGRCLVWDATCVDTVAPCHLDGTSSRPGSAAASAEITKKRKYSGLGSSYSFLPFAVETMGPWGPEAKKFFTEISKRLKEVTHDAKAGWYFAQRVSLAVQRGNVASILGTVPPSSALEDLFYCVG
Summary
Similarity
Belongs to the amiloride-sensitive sodium channel (TC 1.A.6) family.
Uniprot
EMBL
LNIX01000012
OXA48253.1
KQ460779
KPJ12288.1
LNIX01000003
OXA58286.1
+ More
LNIX01000001 OXA61805.1 GEZM01032301 JAV84680.1 LNIX01000038 OXA39608.1 LNIX01000010 OXA49381.1 OXA63637.1 LNIX01000011 OXA48561.1 BDGG01000007 GAV01296.1 BDGG01000002 GAU92213.1 CM007906 OTF86222.1 CM007893 OTG26899.1 CM007892 OTG32916.1 PKPP01001646 PWA80994.1 DF974808 GAU50575.1 CM007897 OTG18487.1 LSRX01000064 OLQ11161.1 ASPP01026421 ETO07174.1 ASPP01007617 ETO26815.1 ASPP01027314 ETO06245.1
LNIX01000001 OXA61805.1 GEZM01032301 JAV84680.1 LNIX01000038 OXA39608.1 LNIX01000010 OXA49381.1 OXA63637.1 LNIX01000011 OXA48561.1 BDGG01000007 GAV01296.1 BDGG01000002 GAU92213.1 CM007906 OTF86222.1 CM007893 OTG26899.1 CM007892 OTG32916.1 PKPP01001646 PWA80994.1 DF974808 GAU50575.1 CM007897 OTG18487.1 LSRX01000064 OLQ11161.1 ASPP01026421 ETO07174.1 ASPP01007617 ETO26815.1 ASPP01027314 ETO06245.1
Proteomes
Pfam
Interpro
IPR036388
WH-like_DNA-bd_sf
+ More
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR000477 RT_dom
IPR019786 Zinc_finger_PHD-type_CS
IPR003150 DNA-bd_RFX
IPR036390 WH_DNA-bd_sf
IPR001873 ENaC
IPR004875 DDE_SF_endonuclease_dom
IPR020903 ENaC_CS
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR004263 Exostosin
IPR026960 RVT-Znf
IPR011011 Znf_FYVE_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR019787 Znf_PHD-finger
IPR000477 RT_dom
IPR019786 Zinc_finger_PHD-type_CS
IPR003150 DNA-bd_RFX
IPR036390 WH_DNA-bd_sf
IPR001873 ENaC
IPR004875 DDE_SF_endonuclease_dom
IPR020903 ENaC_CS
IPR013087 Znf_C2H2_type
IPR036236 Znf_C2H2_sf
IPR004263 Exostosin
IPR026960 RVT-Znf
Gene 3D
ProteinModelPortal
PDB
6AR3
E-value=0.00963101,
Score=95
Ontologies
KEGG
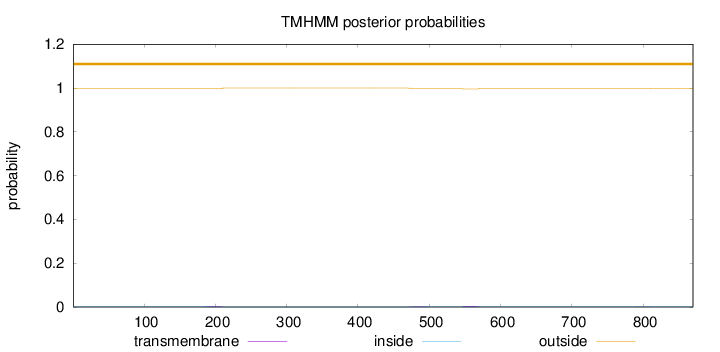
Topology
Length:
870
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.22346
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00371
outside
1 - 870
Population Genetic Test Statistics
Pi
3.052054
Theta
14.191278
Tajima's D
1.733369
CLR
167.181643
CSRT
0.844057797110145
Interpretation
Uncertain
