Gene
KWMTBOMO13678
Annotation
THAP_domain-containing_protein_9?_partial_[Camponotus_floridanus]
Location in the cell
Mitochondrial Reliability : 2.3 Nuclear Reliability : 1.706
Sequence
CDS
ATGCAAATCAGAAGGCTTCAAAAGAAAACCAGCCGTTATCAGAAAAAAATTGCTAATTTAAAGGACATTTTGAAAAACCTTAATGAAAAAAACTTCATTAGTGCTGATCACGTTACGCAATTAGAAAACATTGGAGTAACCGACCTTATCCAGCGTTATGAAAGAAATGTTTCATCGGGTCAAAGTACTAACCAGAAATATCCGCCTGCTTTAAGAATTTTTGCCACTACTCTTCACTTTTATTCGCCAAAGGCGTACGCGTATGTAAGAAAAAAATTCCAAACAGCTTTGCCTCATATTCGAACCATAAAAAAGTGGTACCAATCTATCGATGCTCAGCCTGGATTCACGCAAGAATCATTAAAAGCGATAAAACTTAAAACTGCCTGCACTAAACACAAATTGTTGTGTAATTTAGTTTTAGATGAAATGGCAATCAGACAGCATGTAGAGTGGGATGGAAAACAAATGCACGGGTATGTTGATATTTCTAATTCTGCAGAACAAGGGGATTGTTTACCCGAAGCCAAAGAAGTTCTTGTATTTTTAGTTACCGCAATTAATGGGGCTTGGAAAATACCAGTCGGGTATTTTCTCATTGACGGTGTCACAGGAGAACAACGGTCTAATCTTGTCAGACAATGTTTGGAATTATTAGAAGATACAGGTATTCAAGTTACCTCGATGACTTTTGACGGCTGTCCAGCCAATATCACCATGGCTAAGAAACTCGGATGTTCTTTTGAAGTTGAAAACTTAAAATCATATTTTGAGCACCCCGTTACTCGTCAACCTGTACATATTTTTCTCGATCCATGCCACATGCTGAAGCTATTGAGAAATGCTTTTGAATCATATGGATGTTTCATGGATGCTTCAAATAGGTGTATAAAATGGAGTCATCTAAAAAATCTGTATGACATTCAGGATAAAGAAAAACTGCACTTAGCAAATAAATTACGAAAAAGTCATATCTTTTTCAAGAACCAAAAAATGAAAGTTCGCTTAGCTTCGCAATTATTTAGCAACAGTGTTGCAGATGCCCTGAATTTTTGTTGTGAACTTCAAATCGCTGGTTTTGATGATGTAGAAGGCACTGTGGAATATCTAAAAACTATCAACAACTTGTTCGATGTGTTAAATTCAAGGCATATGGCCCAACTCCATTATAAAAAACCAGTTTGCCCAGGCAACAAAGACAAGGTATTTGAAACTTTAGAAAAGGCGAAACGGTGTCTAACATCTATAAAGCTTTCAGATGGTACAAAAGCTATCATTACACCCAGAAAAACCGGTTTTATTGGTTTATTAGTGTGCATAGAAAGCCTCAAGGCCTTGTATATACAACTAATTGAAAAGGACCAAAAACTTAAGTACATTCCGGCTTACAAAATCAGCCAAGATCATTTAGAGCTTATGTTTGGACATATACGGTCACATGGTGGATGCAGCACCAACCCAACGGCGCGACAGTTTCAAGCAATATATAAAAAGCTCTTGGTAAAGTCGGAGCTTAGAGATGTAGACCAGGGAAACTGTATAGCTCTTGAGGAGGTTTCTTTTTTGTCGTGCTCTTCAGCTGTCCAAAATATAAATCTGACAATCTAA
Protein
MQIRRLQKKTSRYQKKIANLKDILKNLNEKNFISADHVTQLENIGVTDLIQRYERNVSSGQSTNQKYPPALRIFATTLHFYSPKAYAYVRKKFQTALPHIRTIKKWYQSIDAQPGFTQESLKAIKLKTACTKHKLLCNLVLDEMAIRQHVEWDGKQMHGYVDISNSAEQGDCLPEAKEVLVFLVTAINGAWKIPVGYFLIDGVTGEQRSNLVRQCLELLEDTGIQVTSMTFDGCPANITMAKKLGCSFEVENLKSYFEHPVTRQPVHIFLDPCHMLKLLRNAFESYGCFMDASNRCIKWSHLKNLYDIQDKEKLHLANKLRKSHIFFKNQKMKVRLASQLFSNSVADALNFCCELQIAGFDDVEGTVEYLKTINNLFDVLNSRHMAQLHYKKPVCPGNKDKVFETLEKAKRCLTSIKLSDGTKAIITPRKTGFIGLLVCIESLKALYIQLIEKDQKLKYIPAYKISQDHLELMFGHIRSHGGCSTNPTARQFQAIYKKLLVKSELRDVDQGNCIALEEVSFLSCSSAVQNINLTI
Summary
Uniprot
A0A2H1VM77
A0A1Y1JYF3
A0A1Y1JVC6
A0A1Y1K320
E2AQS3
A0A1Y1KU95
+ More
J9KAE9 A0A2R5L6W6 X1X489 X1X5Z2 J9JZH0 A0A1Y1MGQ7 A0A1Y1MGQ8 A0A151IJM3 A0A151IJP2 A0A2A4JUC7 J9M688 A0A3S2PFD8 J9LNJ4 A0A2S2PE11 A0A151WXA5 A0A2S2NIL6 A0A2S2PNR3 J9KWL5 A0A2A4JJZ8 J9KRW2 J9LRN9 J9JRC6 J9L444 A0A1E1XEX7 J9K966 A0A026VY20 A0A146NQI8 A0A1A8B8M0 A0A1A8U049 I3JY54 J9JUB1 J9LWE0 A0A151J7W4 T2M5L7 J9LWZ8 J9K2S6 A0A1A8CVN9 J9LL83 J9LZ19 A0A151J4W4 A0A2S2PEK2 J9LVQ5 A0A1Y1LE13 A0A1A8D9U5 J9KY26 J9JTA4 J9JS62 X1X8L8 J9KRT1 A0A151IXW5 A0A151IM28 E2A8K2 J9M2N4 A0A3S2L852 J9KVX0 A0A1B0DIH9 A0A1Y1NAF9 J9LK29 A0A2G8K6E3 A0A1Y1K3Z7 A0A1Y1NAZ4 A0A1Y1KUU3 A0A2S2NN55 V5G3X1 A0A1Y1KXB8 J9KNF9 A0A151IXT1 A0A151IN16 J9KZX8 A0A1Y1MM65 A0A3N0YYE5 A0A1Y1MGS0 X1XME1 A0A1B0DHA7 A0A2S2PAX0 A0A151IJ13 A0A2S2PS39 J9L4L9 A0A1B0DHV4 A0A151J4S4 J9LU41 X1XT76 A0A151X5G2 J9LVS7 A0A151JB92 J9KLW0 X1WIR4 A0A0J7KTA5 A0A1B0D745
J9KAE9 A0A2R5L6W6 X1X489 X1X5Z2 J9JZH0 A0A1Y1MGQ7 A0A1Y1MGQ8 A0A151IJM3 A0A151IJP2 A0A2A4JUC7 J9M688 A0A3S2PFD8 J9LNJ4 A0A2S2PE11 A0A151WXA5 A0A2S2NIL6 A0A2S2PNR3 J9KWL5 A0A2A4JJZ8 J9KRW2 J9LRN9 J9JRC6 J9L444 A0A1E1XEX7 J9K966 A0A026VY20 A0A146NQI8 A0A1A8B8M0 A0A1A8U049 I3JY54 J9JUB1 J9LWE0 A0A151J7W4 T2M5L7 J9LWZ8 J9K2S6 A0A1A8CVN9 J9LL83 J9LZ19 A0A151J4W4 A0A2S2PEK2 J9LVQ5 A0A1Y1LE13 A0A1A8D9U5 J9KY26 J9JTA4 J9JS62 X1X8L8 J9KRT1 A0A151IXW5 A0A151IM28 E2A8K2 J9M2N4 A0A3S2L852 J9KVX0 A0A1B0DIH9 A0A1Y1NAF9 J9LK29 A0A2G8K6E3 A0A1Y1K3Z7 A0A1Y1NAZ4 A0A1Y1KUU3 A0A2S2NN55 V5G3X1 A0A1Y1KXB8 J9KNF9 A0A151IXT1 A0A151IN16 J9KZX8 A0A1Y1MM65 A0A3N0YYE5 A0A1Y1MGS0 X1XME1 A0A1B0DHA7 A0A2S2PAX0 A0A151IJ13 A0A2S2PS39 J9L4L9 A0A1B0DHV4 A0A151J4S4 J9LU41 X1XT76 A0A151X5G2 J9LVS7 A0A151JB92 J9KLW0 X1WIR4 A0A0J7KTA5 A0A1B0D745
EMBL
ODYU01003321
SOQ41917.1
GEZM01099613
GEZM01099610
GEZM01099604
GEZM01099599
+ More
JAV53268.1 GEZM01099608 GEZM01099605 JAV53264.1 GEZM01099609 JAV53257.1 GL441823 EFN64217.1 GEZM01077470 JAV63295.1 ABLF02011460 ABLF02033413 GGLE01001112 MBY05238.1 ABLF02007004 ABLF02007005 ABLF02008523 ABLF02012848 ABLF02012849 ABLF02012851 GEZM01031190 GEZM01031189 GEZM01031180 JAV84979.1 GEZM01031195 GEZM01031187 GEZM01031186 GEZM01031184 GEZM01031182 GEZM01031181 JAV84992.1 KQ977321 KYN03587.1 KQ977365 KYN03296.1 NWSH01000621 PCG75234.1 ABLF02038040 ABLF02065516 RSAL01000057 RVE49849.1 ABLF02016100 GGMR01014547 MBY27166.1 KQ982672 KYQ52446.1 GGMR01004395 MBY17014.1 GGMR01018395 MBY31014.1 ABLF02016252 NWSH01001223 PCG72048.1 ABLF02011489 ABLF02009662 ABLF02031649 ABLF02031650 GFAC01001507 JAT97681.1 ABLF02016193 KK107641 EZA48371.1 GCES01153042 JAQ33280.1 HADY01024360 SBP62845.1 HAEJ01000494 SBS40951.1 AERX01031390 ABLF02033156 ABLF02003302 KQ979651 KYN20001.1 HAAD01001124 CDG67356.1 ABLF02013367 ABLF02015440 ABLF02057211 ABLF02009087 HADZ01019172 SBP83113.1 ABLF02015114 ABLF02008676 ABLF02008677 KQ980094 KYN17778.1 GGMR01015195 MBY27814.1 ABLF02015299 GEZM01060283 JAV71121.1 HAEA01002407 SBQ30887.1 ABLF02003891 ABLF02003892 ABLF02003893 ABLF02003894 ABLF02014815 ABLF02014817 ABLF02066539 ABLF02019247 ABLF02019254 ABLF02032544 ABLF02032554 ABLF02056709 ABLF02067302 ABLF02042324 ABLF02062307 ABLF02067566 KQ981062 KQ980795 KYN09968.1 KYN13021.1 KQ977081 KYN05929.1 GL437635 EFN70236.1 ABLF02005485 RSAL01000093 RVE47898.1 ABLF02003109 ABLF02024163 ABLF02065890 ABLF02066605 AJVK01034414 GEZM01007799 JAV94911.1 ABLF02018098 MRZV01000842 PIK43542.1 GEZM01095994 JAV55218.1 GEZM01007800 JAV94909.1 GEZM01073324 JAV65152.1 GGMR01005938 MBY18557.1 GALX01003736 JAB64730.1 GEZM01073325 JAV65151.1 ABLF02010172 KQ980298 KQ978748 KYN13019.1 KYN16830.1 KYN28629.1 KQ976993 KYN06573.1 ABLF02017766 GEZM01031193 GEZM01031192 JAV84976.1 RJVU01019282 ROL50904.1 GEZM01031183 JAV84989.1 ABLF02005824 ABLF02005825 ABLF02005826 ABLF02005827 ABLF02050516 ABLF02053731 AJVK01060859 GGMR01013915 MBY26534.1 KQ977417 KYN02849.1 GGMR01019037 MBY31656.1 ABLF02041912 AJVK01061645 KYN17779.1 ABLF02031636 ABLF02031641 ABLF02031645 ABLF02031655 ABLF02046386 ABLF02048949 ABLF02010159 KQ982514 KYQ55617.1 ABLF02008986 ABLF02008987 KQ979155 KYN22412.1 ABLF02005565 ABLF02021063 LBMM01003425 KMQ93551.1 AJVK01026634
JAV53268.1 GEZM01099608 GEZM01099605 JAV53264.1 GEZM01099609 JAV53257.1 GL441823 EFN64217.1 GEZM01077470 JAV63295.1 ABLF02011460 ABLF02033413 GGLE01001112 MBY05238.1 ABLF02007004 ABLF02007005 ABLF02008523 ABLF02012848 ABLF02012849 ABLF02012851 GEZM01031190 GEZM01031189 GEZM01031180 JAV84979.1 GEZM01031195 GEZM01031187 GEZM01031186 GEZM01031184 GEZM01031182 GEZM01031181 JAV84992.1 KQ977321 KYN03587.1 KQ977365 KYN03296.1 NWSH01000621 PCG75234.1 ABLF02038040 ABLF02065516 RSAL01000057 RVE49849.1 ABLF02016100 GGMR01014547 MBY27166.1 KQ982672 KYQ52446.1 GGMR01004395 MBY17014.1 GGMR01018395 MBY31014.1 ABLF02016252 NWSH01001223 PCG72048.1 ABLF02011489 ABLF02009662 ABLF02031649 ABLF02031650 GFAC01001507 JAT97681.1 ABLF02016193 KK107641 EZA48371.1 GCES01153042 JAQ33280.1 HADY01024360 SBP62845.1 HAEJ01000494 SBS40951.1 AERX01031390 ABLF02033156 ABLF02003302 KQ979651 KYN20001.1 HAAD01001124 CDG67356.1 ABLF02013367 ABLF02015440 ABLF02057211 ABLF02009087 HADZ01019172 SBP83113.1 ABLF02015114 ABLF02008676 ABLF02008677 KQ980094 KYN17778.1 GGMR01015195 MBY27814.1 ABLF02015299 GEZM01060283 JAV71121.1 HAEA01002407 SBQ30887.1 ABLF02003891 ABLF02003892 ABLF02003893 ABLF02003894 ABLF02014815 ABLF02014817 ABLF02066539 ABLF02019247 ABLF02019254 ABLF02032544 ABLF02032554 ABLF02056709 ABLF02067302 ABLF02042324 ABLF02062307 ABLF02067566 KQ981062 KQ980795 KYN09968.1 KYN13021.1 KQ977081 KYN05929.1 GL437635 EFN70236.1 ABLF02005485 RSAL01000093 RVE47898.1 ABLF02003109 ABLF02024163 ABLF02065890 ABLF02066605 AJVK01034414 GEZM01007799 JAV94911.1 ABLF02018098 MRZV01000842 PIK43542.1 GEZM01095994 JAV55218.1 GEZM01007800 JAV94909.1 GEZM01073324 JAV65152.1 GGMR01005938 MBY18557.1 GALX01003736 JAB64730.1 GEZM01073325 JAV65151.1 ABLF02010172 KQ980298 KQ978748 KYN13019.1 KYN16830.1 KYN28629.1 KQ976993 KYN06573.1 ABLF02017766 GEZM01031193 GEZM01031192 JAV84976.1 RJVU01019282 ROL50904.1 GEZM01031183 JAV84989.1 ABLF02005824 ABLF02005825 ABLF02005826 ABLF02005827 ABLF02050516 ABLF02053731 AJVK01060859 GGMR01013915 MBY26534.1 KQ977417 KYN02849.1 GGMR01019037 MBY31656.1 ABLF02041912 AJVK01061645 KYN17779.1 ABLF02031636 ABLF02031641 ABLF02031645 ABLF02031655 ABLF02046386 ABLF02048949 ABLF02010159 KQ982514 KYQ55617.1 ABLF02008986 ABLF02008987 KQ979155 KYN22412.1 ABLF02005565 ABLF02021063 LBMM01003425 KMQ93551.1 AJVK01026634
Proteomes
Interpro
Gene 3D
ProteinModelPortal
A0A2H1VM77
A0A1Y1JYF3
A0A1Y1JVC6
A0A1Y1K320
E2AQS3
A0A1Y1KU95
+ More
J9KAE9 A0A2R5L6W6 X1X489 X1X5Z2 J9JZH0 A0A1Y1MGQ7 A0A1Y1MGQ8 A0A151IJM3 A0A151IJP2 A0A2A4JUC7 J9M688 A0A3S2PFD8 J9LNJ4 A0A2S2PE11 A0A151WXA5 A0A2S2NIL6 A0A2S2PNR3 J9KWL5 A0A2A4JJZ8 J9KRW2 J9LRN9 J9JRC6 J9L444 A0A1E1XEX7 J9K966 A0A026VY20 A0A146NQI8 A0A1A8B8M0 A0A1A8U049 I3JY54 J9JUB1 J9LWE0 A0A151J7W4 T2M5L7 J9LWZ8 J9K2S6 A0A1A8CVN9 J9LL83 J9LZ19 A0A151J4W4 A0A2S2PEK2 J9LVQ5 A0A1Y1LE13 A0A1A8D9U5 J9KY26 J9JTA4 J9JS62 X1X8L8 J9KRT1 A0A151IXW5 A0A151IM28 E2A8K2 J9M2N4 A0A3S2L852 J9KVX0 A0A1B0DIH9 A0A1Y1NAF9 J9LK29 A0A2G8K6E3 A0A1Y1K3Z7 A0A1Y1NAZ4 A0A1Y1KUU3 A0A2S2NN55 V5G3X1 A0A1Y1KXB8 J9KNF9 A0A151IXT1 A0A151IN16 J9KZX8 A0A1Y1MM65 A0A3N0YYE5 A0A1Y1MGS0 X1XME1 A0A1B0DHA7 A0A2S2PAX0 A0A151IJ13 A0A2S2PS39 J9L4L9 A0A1B0DHV4 A0A151J4S4 J9LU41 X1XT76 A0A151X5G2 J9LVS7 A0A151JB92 J9KLW0 X1WIR4 A0A0J7KTA5 A0A1B0D745
J9KAE9 A0A2R5L6W6 X1X489 X1X5Z2 J9JZH0 A0A1Y1MGQ7 A0A1Y1MGQ8 A0A151IJM3 A0A151IJP2 A0A2A4JUC7 J9M688 A0A3S2PFD8 J9LNJ4 A0A2S2PE11 A0A151WXA5 A0A2S2NIL6 A0A2S2PNR3 J9KWL5 A0A2A4JJZ8 J9KRW2 J9LRN9 J9JRC6 J9L444 A0A1E1XEX7 J9K966 A0A026VY20 A0A146NQI8 A0A1A8B8M0 A0A1A8U049 I3JY54 J9JUB1 J9LWE0 A0A151J7W4 T2M5L7 J9LWZ8 J9K2S6 A0A1A8CVN9 J9LL83 J9LZ19 A0A151J4W4 A0A2S2PEK2 J9LVQ5 A0A1Y1LE13 A0A1A8D9U5 J9KY26 J9JTA4 J9JS62 X1X8L8 J9KRT1 A0A151IXW5 A0A151IM28 E2A8K2 J9M2N4 A0A3S2L852 J9KVX0 A0A1B0DIH9 A0A1Y1NAF9 J9LK29 A0A2G8K6E3 A0A1Y1K3Z7 A0A1Y1NAZ4 A0A1Y1KUU3 A0A2S2NN55 V5G3X1 A0A1Y1KXB8 J9KNF9 A0A151IXT1 A0A151IN16 J9KZX8 A0A1Y1MM65 A0A3N0YYE5 A0A1Y1MGS0 X1XME1 A0A1B0DHA7 A0A2S2PAX0 A0A151IJ13 A0A2S2PS39 J9L4L9 A0A1B0DHV4 A0A151J4S4 J9LU41 X1XT76 A0A151X5G2 J9LVS7 A0A151JB92 J9KLW0 X1WIR4 A0A0J7KTA5 A0A1B0D745
Ontologies
GO
PANTHER
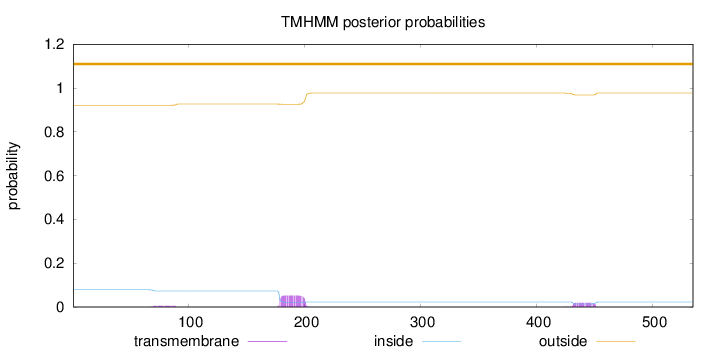
Topology
Length:
535
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.61461
Exp number, first 60 AAs:
0
Total prob of N-in:
0.07833
outside
1 - 535
Population Genetic Test Statistics
Pi
112.318873
Theta
146.84106
Tajima's D
-0.906712
CLR
75.437796
CSRT
0.152842357882106
Interpretation
Uncertain
