Gene
KWMTBOMO13675
Pre Gene Modal
BGIBMGA011082
Annotation
PREDICTED:_slit_homolog_3_protein-like_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.968
Sequence
CDS
ATGTGGTGCTGGACATGTCAGGGTGCCGCTGCGGCGTCACTGACAAGTAGAGTGGCGGAGGCGCCACAGGAGTGTGAATGGCAGCGGGTTTCTGGCGATGCAAGCGAGAGTACCAGAGTGCAGCTCGCGTGCTCCTTGAGGACCGCAGCCGGAGCGACTGAGCTACTGACCGGTCTGAGTGCTTCACAGGCTCAACGCATCACAGTTCTAGACCTTCATTGTACCGATGTTCTCTTTTTTGAGAGTTCACTTGATTTTGGTCGAAGAAAAGAAGAGGGAACTCAATTGCTGTCCCGGTTTCACAACTTGAGAGAATTAAGAATAGAATCCTGTAAAATACGCTATGTGCCGTCAGCGGTGCTATCACCGCTTAGTGGCCTACGTAGTTTGACTATTCGGACTCATAATTCTGATTGGTCAGCGATGTCGATGGAATTCCACAGAGATACTTTTCGAGGGCTGACGGATTTGAGATCACTTGATTTAGGCGATAACAACATATGGATTTTGCCGTCGGAAATATTTTGTGTTCTGTACAATTTGAAAGAACTAAATGTTACCCAGAATAGATTGCAAGATATATCAAACTTAGGCTTTTCTGACTGGGGTAATGGACCAACGGCACCTGGTAAATCGTGTAACACAGTTCTTGAGACCTTGGATATGTCGTACAATGAAATTAGCGGCCTACCTGATAACGGTTTGTCAAGTTTACGCGCACTTCAAAAATTGTTTCTGCAAAACAATAGGATTTCAAACGTCGCTGACCGAGCTTTCGTAGGTTTAAGTGACTTACAAATACTGAATTTATCAACAAATGCTCTAACTGCCTTGCCTCCGGAGATGTTTCAATCTTCAAGAGATATAAAACAAATTTACTTAAACAACAACAGCTTAAGCGTATTGGCGCCTGGACTTCTAGAAGGATTAGACCAATTGCAGATATTAGATTTATCAGTCAATGAATTGACTAGTGAATGGATAAACAGAGATACATTCTCTGGATTGGTTCGGTTGATTGTTCTGAATCTATCTCACAACAGAATTACGAAAATTGATGCTCTATTATTTCAAGATCTTAACAACCTACAATTTCTGAGTTTGGAATATAATAACGTAGGTCGTATCGCAGAAGGTGCATTTTCGAACTTGAAGAATCTTCATTCCCTTTCACTGGCTCACAATAACATCATAGAAGTGGATAGCGGTCATTTCTCAAATCTATACGTATTAAATCAGCTGTTTCTTGACGGCAACAGAATTACTAAGGTCGACATAAGGTCTTTTGAAAATATTACAAAACTCCACGACTTGGGTTTGAGTGGAAATCAACTAACCGACGTACCTGAAGCGATCAAGACTTTAAGGTTCTTAACATCCTTGGATTTAGGAATGAATAAAATAACAAAAGTGATAACTTCGCACTTTGACGGCCTTGATGATTTGTATGGTTTAAGACTTGTAGGAAATAAAATCGAGAAAATTTCTAAGGATACATTTGCGGCTTTACCGTCTCTGCAAATATTGAACTTGGCTTCGAATAACATCGATCAAATAGACGACGGGGCTTTTGCGTCGAACCAGCAGTTGAAAGCGATTGGATTAGATGGTAACAAATTGGTAGACTTAAAAGGGATTTTTACTAAAACTCAGCCGCTTGTCTGGTTAAATGTATCCAACAATGAATTACTTTGGTTTGATTACAGTCATATACCATCCAATCTAGAATGGTTAGATATGCACGAAAATAAAATTGACAAGTTAGATGATACGTATGGATTGAAGGAGACAACCATGGTGAAGATGCTCGATGTTAGTAATAACAAAATCAAGAGCGTAGATGAGCTTTCCTTTCCAAGCAGTATTGAGACTGTAGTTTTAAACAATAATAATATTGAAAAAGTTAGTCCCGGTACATTCTTGCAAAAATACAATTTGAATAAGGTTATGTTGTATTCGAACAAAATAAAAACATTAGAGCTCGGTGCATTCGCGATATCATCTGTTCCAGACGATAAAGATTTACCAGAGTTCTATATCAGTGAAAATCCTTTTGTGTGCGACTGCACAATGGAATGGCTCCAAACAATTAATCAAGTAAGCGACCTCAGGCAGCGCCCAAGAGTAATGGATTTAGAAAGCGTGAGGTGCTCTTTAACCCATTCACGGAGGAAATCGGAGACGCTCTTGCTCGAAGTGAAGTCTTCAGAATTTTTGTGCGAATATGAATCGCACTGCTTCACAGTCTGTCATTGCTGTGACTTTGACGCCTGCGATTGTAAGATGACCTGTCCAGATAGGTGTTCGTGCTATCACGATCAGACTTGGACCACCACTAACGTAGTCGATTGTTCGAATGCCGGCTACGATCACGTGCCCGACAGAATACCGATGGACGCGACCGAAATATATTTAGATGGTAACAGTTTGAAAGAGCTCGGAAATCACGTTTTCATTGGCAAAAAGAAACTACAAGTTTTATATCTCAACAACAGTAATATTGATATAATACAAAATAGAACATTTAACGGAATCGACTCTTTGAGAGTGCTCCATTTAGAAAACAATAACTTGGAAGTATTGAGGAACACGCAATTCACGAGGCTACCTCATTTGAACGAGCTCTACTTGAGCAGCAATAAGATCAAATTTATCGAGAATGATACTTTTAATTATCTGCCTTCTTTAGAGTTCTTGGACCTCGATAATAATGGTTACGTTGATTACATGCCGTGGCGAGTCATTTCCGACAGCGATCCGCGCATGCGTGTCTCTGTAGATGGAAACAACTGGATATGTGACTGTAAAGATGTGGCTCAATTGAATCAGTGGCTAATAAAAAAATCAAAAGACACCGAGAACATGATGTGCTACTTTGCTCACGGTCAGCCCATGAATAAAACCATCGCGACCGTTGCTAAGGAATGTAAAACAGAAACGACTACCGAAGAAACTGGTACCGAGACGTTAAAACGATTATTCATCGAATCGAATGACGGCGTCGAAAATTATATTCCATATATTGCAGCCGTGTTGATAATAGCAATCATTCTATTACTGATGTGCGCTCTTCTTTTCATGTTCAGAGAAGATTTTAAACTATGGATGCACTCTAGATATGGTATTAGAATATTCAGTTCCTCCCCCCGGGACCTTAACGACAGCAAAAACAAACGGTTTGATGCATTTTTTGTGTATAATCCACGCGACGAAGACTTTGTGACTCGGGCGGTGAGTTCGGAATTAGAAAATATGGGACACACTTTGTGCTTACAACACCGGGACCTGCAGCTTATAGAGAGACGTTCGGGTGATAGTTTAGTGAGTGCCGCGGAAAGTTCTAAGAGACTGATAATAGTTTTATCGATAAACTTTTTACAACAAGAGTGGTACGCGCCTGAATCTAAAGCGGCGGTGCAAAGTGCTATAAATTCAGTGAATGTGAGACACAGACGACAAAAGATAATTTTTCTAGTAACAACAGATTTAAGTGCCATAAACATAGATCCTGACTTGAAAGTGCTTCTTAAAACGTGTACAGTGATCGTGTGGGGTGAGAGAAATTGTTGGGAAAAACTAAATTTTCGGTTACCCGATGTGGACGTAACGTTACCGAATCGGACGCTTCATAACGCTAATAATATTATAAAAAGTGGTAACAGAGAAGGATTCGGTAGGCACGGTAATTTAAGGTACACCGCGCCACCGACTTCTCATGATCCTTGGTACAAATATGGTATGGCTCCTCCAATACTAATGATGTCTAGTCCGTTGCATTCGACCAGCGCCAGCGCGGAAGTCTCTGCTCGGAGCACGGAGGACGAGACGTGTTCGGTCGCTAGCAGTGAAGGTCGACCTGACGACCGTCTGCCTCACCACCATAGCTACGTCTCTATAGACAACCATCAGTGCGAGGAACGCCCTCTTAGGCCCGGCCAGCAAATACCAATGTCCAGTACAGCTCATACAAGAAAGACATATTTCGTTTAA
Protein
MWCWTCQGAAAASLTSRVAEAPQECEWQRVSGDASESTRVQLACSLRTAAGATELLTGLSASQAQRITVLDLHCTDVLFFESSLDFGRRKEEGTQLLSRFHNLRELRIESCKIRYVPSAVLSPLSGLRSLTIRTHNSDWSAMSMEFHRDTFRGLTDLRSLDLGDNNIWILPSEIFCVLYNLKELNVTQNRLQDISNLGFSDWGNGPTAPGKSCNTVLETLDMSYNEISGLPDNGLSSLRALQKLFLQNNRISNVADRAFVGLSDLQILNLSTNALTALPPEMFQSSRDIKQIYLNNNSLSVLAPGLLEGLDQLQILDLSVNELTSEWINRDTFSGLVRLIVLNLSHNRITKIDALLFQDLNNLQFLSLEYNNVGRIAEGAFSNLKNLHSLSLAHNNIIEVDSGHFSNLYVLNQLFLDGNRITKVDIRSFENITKLHDLGLSGNQLTDVPEAIKTLRFLTSLDLGMNKITKVITSHFDGLDDLYGLRLVGNKIEKISKDTFAALPSLQILNLASNNIDQIDDGAFASNQQLKAIGLDGNKLVDLKGIFTKTQPLVWLNVSNNELLWFDYSHIPSNLEWLDMHENKIDKLDDTYGLKETTMVKMLDVSNNKIKSVDELSFPSSIETVVLNNNNIEKVSPGTFLQKYNLNKVMLYSNKIKTLELGAFAISSVPDDKDLPEFYISENPFVCDCTMEWLQTINQVSDLRQRPRVMDLESVRCSLTHSRRKSETLLLEVKSSEFLCEYESHCFTVCHCCDFDACDCKMTCPDRCSCYHDQTWTTTNVVDCSNAGYDHVPDRIPMDATEIYLDGNSLKELGNHVFIGKKKLQVLYLNNSNIDIIQNRTFNGIDSLRVLHLENNNLEVLRNTQFTRLPHLNELYLSSNKIKFIENDTFNYLPSLEFLDLDNNGYVDYMPWRVISDSDPRMRVSVDGNNWICDCKDVAQLNQWLIKKSKDTENMMCYFAHGQPMNKTIATVAKECKTETTTEETGTETLKRLFIESNDGVENYIPYIAAVLIIAIILLLMCALLFMFREDFKLWMHSRYGIRIFSSSPRDLNDSKNKRFDAFFVYNPRDEDFVTRAVSSELENMGHTLCLQHRDLQLIERRSGDSLVSAAESSKRLIIVLSINFLQQEWYAPESKAAVQSAINSVNVRHRRQKIIFLVTTDLSAINIDPDLKVLLKTCTVIVWGERNCWEKLNFRLPDVDVTLPNRTLHNANNIIKSGNREGFGRHGNLRYTAPPTSHDPWYKYGMAPPILMMSSPLHSTSASAEVSARSTEDETCSVASSEGRPDDRLPHHHSYVSIDNHQCEERPLRPGQQIPMSSTAHTRKTYFV
Summary
Similarity
Belongs to the Toll-like receptor family.
Uniprot
A0A2A4JYQ2
A0A2H1VLQ9
A0A346RAF7
A0A0L7L0F7
A0A194REQ7
A0A194PL06
+ More
H9JNH9 A0A212EVE6 A0A2P8YLA0 A0A2J7QTB8 A0A067RDD5 T1HTM0 A0A0A9ZIG4 A0A1B6D9Z8 A0A1Y1N5R0 A0A173ADT7 D6WCH9 E0VC85 A0A336MA92 A0A336LWU3 U4U980 A0A182GFV2 Q17DZ2 A0A194PLB4 A0A194R9V1 N0A0T6 J9K5Y3 A0A2A4JHX4 A0A2S2PI41 A0A2H8TSA7 A0A2H1VEQ6 A0A0L7LDH5 A0A346RAF8 A0A2S2QZS4 A0A084WCF6 A0A182FKH9 W5JR01 A0A182WDG0 A0A182RTF7 A0A182N2E6 A0A182KEJ4 A0A182ML25 A0A182YPU4 A0A182QP16 A0A182NZJ4 A0A182HWF3 A0A182UR06 A0A182LHS2 Q7QHH4 A0A182X5J7 A0A182TLV4 A0A182TJZ3 B0X372 F4WCN0 A0A158NHZ4 A0A195BXM2 A0A195DF99 A0A151WXS3 A0A154NZK5 A0A084WCI0 A0A182H5C2 Q16VI7 A0A195CVE8 A0A0M9A8F0 A0A026WSN2 A0A182YPU6 A0A1S4K220 A0A0L7QT80 Q7QHH1 A0A0C9QMR2 A0A1S4FMM1 A0A088AAK1 A0A182JKN8 A0A0J7KUE8 E2BUC9 H9JNC2 B0XFP7 A0A182X5J9 A0A1S4H5Y8 A0A182UWU4 A0A182WDG2 A0A182K0G0 A0A182NZJ6 A0A182HWF1 A0A182LHS4 A0A182UF17 A0A182MNH4 A0A182RTF5 A0A182Q9N9 A0A182N2E1 E1ZY04 K7JAV1 A0A336LW85 A0A336N8Q7 A0A195ESS7 A0A182JLH8 A0A194R9A1 N6T878 A0A1Y1LF69
H9JNH9 A0A212EVE6 A0A2P8YLA0 A0A2J7QTB8 A0A067RDD5 T1HTM0 A0A0A9ZIG4 A0A1B6D9Z8 A0A1Y1N5R0 A0A173ADT7 D6WCH9 E0VC85 A0A336MA92 A0A336LWU3 U4U980 A0A182GFV2 Q17DZ2 A0A194PLB4 A0A194R9V1 N0A0T6 J9K5Y3 A0A2A4JHX4 A0A2S2PI41 A0A2H8TSA7 A0A2H1VEQ6 A0A0L7LDH5 A0A346RAF8 A0A2S2QZS4 A0A084WCF6 A0A182FKH9 W5JR01 A0A182WDG0 A0A182RTF7 A0A182N2E6 A0A182KEJ4 A0A182ML25 A0A182YPU4 A0A182QP16 A0A182NZJ4 A0A182HWF3 A0A182UR06 A0A182LHS2 Q7QHH4 A0A182X5J7 A0A182TLV4 A0A182TJZ3 B0X372 F4WCN0 A0A158NHZ4 A0A195BXM2 A0A195DF99 A0A151WXS3 A0A154NZK5 A0A084WCI0 A0A182H5C2 Q16VI7 A0A195CVE8 A0A0M9A8F0 A0A026WSN2 A0A182YPU6 A0A1S4K220 A0A0L7QT80 Q7QHH1 A0A0C9QMR2 A0A1S4FMM1 A0A088AAK1 A0A182JKN8 A0A0J7KUE8 E2BUC9 H9JNC2 B0XFP7 A0A182X5J9 A0A1S4H5Y8 A0A182UWU4 A0A182WDG2 A0A182K0G0 A0A182NZJ6 A0A182HWF1 A0A182LHS4 A0A182UF17 A0A182MNH4 A0A182RTF5 A0A182Q9N9 A0A182N2E1 E1ZY04 K7JAV1 A0A336LW85 A0A336N8Q7 A0A195ESS7 A0A182JLH8 A0A194R9A1 N6T878 A0A1Y1LF69
Pubmed
EMBL
NWSH01000423
PCG76502.1
ODYU01003249
SOQ41753.1
MH243023
AXS59157.1
+ More
JTDY01003948 KOB68796.1 KQ460473 KPJ14416.1 KQ459600 KPI94116.1 BABH01019465 AGBW02012218 OWR45424.1 PYGN01000514 PSN45021.1 NEVH01011197 PNF31826.1 KK852574 KDR21046.1 ACPB03003065 GBHO01000879 JAG42725.1 GEDC01030532 GEDC01029683 GEDC01020757 GEDC01014780 GEDC01000406 JAS06766.1 JAS07615.1 JAS16541.1 JAS22518.1 JAS36892.1 GEZM01012073 JAV93233.1 KX086265 KK854633 ANG08888.1 PTY17485.1 KQ971311 EEZ99355.1 DS235049 EEB10991.1 UFQT01000297 SSX22958.1 UFQT01000175 SSX21133.1 KB632184 ERL89622.1 JXUM01060447 KQ562102 KXJ76681.1 CH477289 EAT44679.1 KPI94117.1 KPJ14417.1 KC355238 AGK40938.1 ABLF02037547 NWSH01001355 PCG71561.1 GGMR01016492 MBY29111.1 GFXV01005290 MBW17095.1 ODYU01002158 SOQ39297.1 JTDY01001660 KOB73226.1 MH243024 AXS59158.1 GGMS01014055 MBY83258.1 ATLV01022666 KE525335 KFB47900.1 ADMH02000685 ETN65330.1 AXCM01004773 AXCN02001726 APCN01001487 AAAB01008816 EAA05150.4 DS232310 EDS39574.1 GL888070 EGI68210.1 ADTU01001755 KQ976398 KYM92696.1 KQ980905 KYN11517.1 KQ982662 KYQ52653.1 KQ434786 KZC05057.1 ATLV01022669 KFB47924.1 JXUM01111276 KQ565488 KXJ70919.1 CH477591 EAT38569.2 KQ977259 KYN04610.1 KQ435727 KOX78043.1 KK107111 QOIP01000011 EZA59042.1 RLU16722.1 KQ414755 KOC61774.1 EAA05071.4 GBYB01004774 JAG74541.1 LBMM01003120 KMQ93943.1 GL450630 EFN80713.1 BABH01019497 DS232948 EDS26928.1 APCN01001486 AXCM01015215 AXCN02001727 GL435158 EFN73944.1 AAZX01003450 AAZX01009988 UFQT01000128 SSX20587.1 UFQS01003570 UFQT01003570 SSX15766.1 SSX35118.1 KQ981979 KYN31305.1 KPJ14418.1 APGK01047889 KB741092 KB632050 ENN73923.1 ERL88314.1 GEZM01061234 JAV70615.1
JTDY01003948 KOB68796.1 KQ460473 KPJ14416.1 KQ459600 KPI94116.1 BABH01019465 AGBW02012218 OWR45424.1 PYGN01000514 PSN45021.1 NEVH01011197 PNF31826.1 KK852574 KDR21046.1 ACPB03003065 GBHO01000879 JAG42725.1 GEDC01030532 GEDC01029683 GEDC01020757 GEDC01014780 GEDC01000406 JAS06766.1 JAS07615.1 JAS16541.1 JAS22518.1 JAS36892.1 GEZM01012073 JAV93233.1 KX086265 KK854633 ANG08888.1 PTY17485.1 KQ971311 EEZ99355.1 DS235049 EEB10991.1 UFQT01000297 SSX22958.1 UFQT01000175 SSX21133.1 KB632184 ERL89622.1 JXUM01060447 KQ562102 KXJ76681.1 CH477289 EAT44679.1 KPI94117.1 KPJ14417.1 KC355238 AGK40938.1 ABLF02037547 NWSH01001355 PCG71561.1 GGMR01016492 MBY29111.1 GFXV01005290 MBW17095.1 ODYU01002158 SOQ39297.1 JTDY01001660 KOB73226.1 MH243024 AXS59158.1 GGMS01014055 MBY83258.1 ATLV01022666 KE525335 KFB47900.1 ADMH02000685 ETN65330.1 AXCM01004773 AXCN02001726 APCN01001487 AAAB01008816 EAA05150.4 DS232310 EDS39574.1 GL888070 EGI68210.1 ADTU01001755 KQ976398 KYM92696.1 KQ980905 KYN11517.1 KQ982662 KYQ52653.1 KQ434786 KZC05057.1 ATLV01022669 KFB47924.1 JXUM01111276 KQ565488 KXJ70919.1 CH477591 EAT38569.2 KQ977259 KYN04610.1 KQ435727 KOX78043.1 KK107111 QOIP01000011 EZA59042.1 RLU16722.1 KQ414755 KOC61774.1 EAA05071.4 GBYB01004774 JAG74541.1 LBMM01003120 KMQ93943.1 GL450630 EFN80713.1 BABH01019497 DS232948 EDS26928.1 APCN01001486 AXCM01015215 AXCN02001727 GL435158 EFN73944.1 AAZX01003450 AAZX01009988 UFQT01000128 SSX20587.1 UFQS01003570 UFQT01003570 SSX15766.1 SSX35118.1 KQ981979 KYN31305.1 KPJ14418.1 APGK01047889 KB741092 KB632050 ENN73923.1 ERL88314.1 GEZM01061234 JAV70615.1
Proteomes
UP000218220
UP000037510
UP000053240
UP000053268
UP000005204
UP000007151
+ More
UP000245037 UP000235965 UP000027135 UP000015103 UP000007266 UP000009046 UP000030742 UP000069940 UP000249989 UP000008820 UP000007819 UP000030765 UP000069272 UP000000673 UP000075920 UP000075900 UP000075884 UP000075881 UP000075883 UP000076408 UP000075886 UP000075885 UP000075840 UP000075903 UP000075882 UP000007062 UP000076407 UP000075902 UP000002320 UP000007755 UP000005205 UP000078540 UP000078492 UP000075809 UP000076502 UP000078542 UP000053105 UP000053097 UP000279307 UP000053825 UP000005203 UP000075880 UP000036403 UP000008237 UP000000311 UP000002358 UP000078541 UP000019118
UP000245037 UP000235965 UP000027135 UP000015103 UP000007266 UP000009046 UP000030742 UP000069940 UP000249989 UP000008820 UP000007819 UP000030765 UP000069272 UP000000673 UP000075920 UP000075900 UP000075884 UP000075881 UP000075883 UP000076408 UP000075886 UP000075885 UP000075840 UP000075903 UP000075882 UP000007062 UP000076407 UP000075902 UP000002320 UP000007755 UP000005205 UP000078540 UP000078492 UP000075809 UP000076502 UP000078542 UP000053105 UP000053097 UP000279307 UP000053825 UP000005203 UP000075880 UP000036403 UP000008237 UP000000311 UP000002358 UP000078541 UP000019118
Interpro
SUPFAM
SSF52200
SSF52200
Gene 3D
ProteinModelPortal
A0A2A4JYQ2
A0A2H1VLQ9
A0A346RAF7
A0A0L7L0F7
A0A194REQ7
A0A194PL06
+ More
H9JNH9 A0A212EVE6 A0A2P8YLA0 A0A2J7QTB8 A0A067RDD5 T1HTM0 A0A0A9ZIG4 A0A1B6D9Z8 A0A1Y1N5R0 A0A173ADT7 D6WCH9 E0VC85 A0A336MA92 A0A336LWU3 U4U980 A0A182GFV2 Q17DZ2 A0A194PLB4 A0A194R9V1 N0A0T6 J9K5Y3 A0A2A4JHX4 A0A2S2PI41 A0A2H8TSA7 A0A2H1VEQ6 A0A0L7LDH5 A0A346RAF8 A0A2S2QZS4 A0A084WCF6 A0A182FKH9 W5JR01 A0A182WDG0 A0A182RTF7 A0A182N2E6 A0A182KEJ4 A0A182ML25 A0A182YPU4 A0A182QP16 A0A182NZJ4 A0A182HWF3 A0A182UR06 A0A182LHS2 Q7QHH4 A0A182X5J7 A0A182TLV4 A0A182TJZ3 B0X372 F4WCN0 A0A158NHZ4 A0A195BXM2 A0A195DF99 A0A151WXS3 A0A154NZK5 A0A084WCI0 A0A182H5C2 Q16VI7 A0A195CVE8 A0A0M9A8F0 A0A026WSN2 A0A182YPU6 A0A1S4K220 A0A0L7QT80 Q7QHH1 A0A0C9QMR2 A0A1S4FMM1 A0A088AAK1 A0A182JKN8 A0A0J7KUE8 E2BUC9 H9JNC2 B0XFP7 A0A182X5J9 A0A1S4H5Y8 A0A182UWU4 A0A182WDG2 A0A182K0G0 A0A182NZJ6 A0A182HWF1 A0A182LHS4 A0A182UF17 A0A182MNH4 A0A182RTF5 A0A182Q9N9 A0A182N2E1 E1ZY04 K7JAV1 A0A336LW85 A0A336N8Q7 A0A195ESS7 A0A182JLH8 A0A194R9A1 N6T878 A0A1Y1LF69
H9JNH9 A0A212EVE6 A0A2P8YLA0 A0A2J7QTB8 A0A067RDD5 T1HTM0 A0A0A9ZIG4 A0A1B6D9Z8 A0A1Y1N5R0 A0A173ADT7 D6WCH9 E0VC85 A0A336MA92 A0A336LWU3 U4U980 A0A182GFV2 Q17DZ2 A0A194PLB4 A0A194R9V1 N0A0T6 J9K5Y3 A0A2A4JHX4 A0A2S2PI41 A0A2H8TSA7 A0A2H1VEQ6 A0A0L7LDH5 A0A346RAF8 A0A2S2QZS4 A0A084WCF6 A0A182FKH9 W5JR01 A0A182WDG0 A0A182RTF7 A0A182N2E6 A0A182KEJ4 A0A182ML25 A0A182YPU4 A0A182QP16 A0A182NZJ4 A0A182HWF3 A0A182UR06 A0A182LHS2 Q7QHH4 A0A182X5J7 A0A182TLV4 A0A182TJZ3 B0X372 F4WCN0 A0A158NHZ4 A0A195BXM2 A0A195DF99 A0A151WXS3 A0A154NZK5 A0A084WCI0 A0A182H5C2 Q16VI7 A0A195CVE8 A0A0M9A8F0 A0A026WSN2 A0A182YPU6 A0A1S4K220 A0A0L7QT80 Q7QHH1 A0A0C9QMR2 A0A1S4FMM1 A0A088AAK1 A0A182JKN8 A0A0J7KUE8 E2BUC9 H9JNC2 B0XFP7 A0A182X5J9 A0A1S4H5Y8 A0A182UWU4 A0A182WDG2 A0A182K0G0 A0A182NZJ6 A0A182HWF1 A0A182LHS4 A0A182UF17 A0A182MNH4 A0A182RTF5 A0A182Q9N9 A0A182N2E1 E1ZY04 K7JAV1 A0A336LW85 A0A336N8Q7 A0A195ESS7 A0A182JLH8 A0A194R9A1 N6T878 A0A1Y1LF69
PDB
4U7L
E-value=1.09083e-34,
Score=372
Ontologies
GO
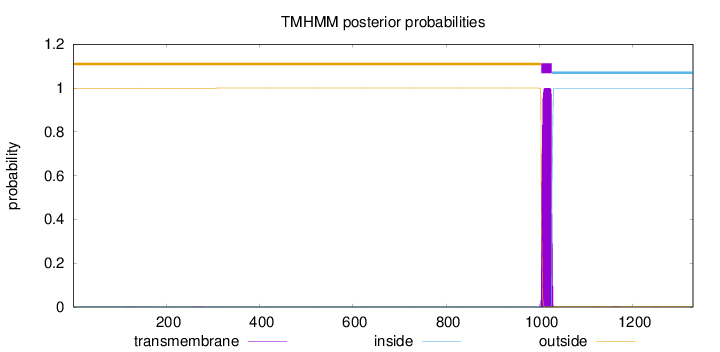
Topology
Length:
1329
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.89126
Exp number, first 60 AAs:
0.00204
Total prob of N-in:
0.00101
outside
1 - 1003
TMhelix
1004 - 1026
inside
1027 - 1329
Population Genetic Test Statistics
Pi
193.330024
Theta
215.376291
Tajima's D
0.036196
CLR
0.234813
CSRT
0.377731113444328
Interpretation
Uncertain
