Gene
KWMTBOMO13672
Pre Gene Modal
BGIBMGA011028
Annotation
PREDICTED:_cell_division_cycle_protein_20_homolog_[Papilio_xuthus]
Location in the cell
Nuclear Reliability : 1.906
Sequence
CDS
ATGGATGGCCCTATAACCCGAGGTCCATTGCAACGGTGGGCTAAAAAAGGCTGTAATGAAAATATAAATCCGTCTCTCAGTAACTCTTTCAAGAATCATTCAAATATTAACATAAGTTCGTGTAATCAATCGCTGCAGAAACTGTCCATATCTGCTAATCAAAGTTGTAACAATAGTGCATTGTCCAATAATAAAACACCAACGAGAAATGAAAATAAGAAAGGAAAAAAGACTCCTTCGAAAACGAAGTCTCCAGGTCATTCAACAACACCAACCCCAAACAAAGCATCAAAGACACCTAATAAAGCTGATAGATTCATCCCATCAAGGAGTAATTCAAATTATGACCTATGCCACTATATGTTGAGTCGTGATGATGACAAAAGTGAGGAAACCCCACAAACAGCTGCCGGAGAAGCAATAGGTAGAGCTCTGAATGATACTGAACCAGGTCGCCTCCTCCAGTATACTTGTAAAGCACCAGCAGCACCCGAAGGCTACCAGAACAGACTTAGAGTAGTATATTCACAGACAAAAGTTCCGTCATCAGTAAAATGCAACACTAGATACATACCTCAGGCTCCTGACAGGATACTTGATGCTCCTGAAATTTTAGATGACTATTATTTGAACCTTGTCGATTGGAGTGCATCAAACATTCTTGCAGTAGCATTAGGAAACTCTGTATATCTATGGAATGCTGGCACGGGACAAATAGAACAACTTCTAACCCTTGAAGGAGCAGAGACAGTCTGTTCTGTGGGATGGGTGCAAGGTGGTGGATCGCATCTGGCTGTTGGGACTTCAATGGCAACTGTAGAATTGTGGGACTGTGAAAAGATCAAACGTTTGAGGGTAATGGATGGTCACAGCGGTCGTGTCGGTTCTCTGGCTTGGAACATGTACATAGTGTCAAGTGGTGCCAGAGATGGACACATTGTTCACCACGACGTCAGGCAAAGAGATCATGCTGTCGCCAACATACATGGACATACACAAGAGATCTGCGGACTGAAATGGTCACCTGACGGTAGATATCTTGCATCGGGCGGCAACGATAATCTGCTGAACATTTGGCCAATTGCACAAGGACAACATTATTCACAGCCACAACATATACTTTCATTCAATCAACATCTAGCCGCAGTGAAAGGTCTCGCGTGGTGTCCGTGGAGCGCCGGCATCCTCGCGTCCGGCGGCGGTACGGCCGACAGAACTATACGGTTATGGAACATCAGTACTGGATCCAATCTTAATACTGTTGACACTAAATCTCAGGTTTGCTCTATAGTATGGAGCACTCACTACAAGGAGCTTATCTCGGGTCATGGGTATGCTCATAACCAACTAATCATATGGAAGTATCCAACAATGTCAAGAGTTGCAGAATTGTCAGGACATATGGCCAGAGTACTGCACCTTGCATTATCGCCTGATGGTACTACAGTACTATCTGCTGGCGCTGACGAAACCCTAAGATTATGGAAGTGCTTTATGTTAGATCCATCAAAGAAAAAAGAACCTACTGACGCGAAGGCAGCTAAGTCGCTGCTGAATCTCAATTCTCTCATCAGATAG
Protein
MDGPITRGPLQRWAKKGCNENINPSLSNSFKNHSNINISSCNQSLQKLSISANQSCNNSALSNNKTPTRNENKKGKKTPSKTKSPGHSTTPTPNKASKTPNKADRFIPSRSNSNYDLCHYMLSRDDDKSEETPQTAAGEAIGRALNDTEPGRLLQYTCKAPAAPEGYQNRLRVVYSQTKVPSSVKCNTRYIPQAPDRILDAPEILDDYYLNLVDWSASNILAVALGNSVYLWNAGTGQIEQLLTLEGAETVCSVGWVQGGGSHLAVGTSMATVELWDCEKIKRLRVMDGHSGRVGSLAWNMYIVSSGARDGHIVHHDVRQRDHAVANIHGHTQEICGLKWSPDGRYLASGGNDNLLNIWPIAQGQHYSQPQHILSFNQHLAAVKGLAWCPWSAGILASGGGTADRTIRLWNISTGSNLNTVDTKSQVCSIVWSTHYKELISGHGYAHNQLIIWKYPTMSRVAELSGHMARVLHLALSPDGTTVLSAGADETLRLWKCFMLDPSKKKEPTDAKAAKSLLNLNSLIR
Summary
Uniprot
A0A2A4JXH4
A0A2H1VW01
A0A3S2NZY5
H9JNC5
A0A212EVB1
S4PDU2
+ More
A0A194RA82 A0A1B6GJ63 A0A1Y1LIH0 K7ISD3 A0A232F777 A0A026WIT0 A0A1B6CMN1 A0A1B6KWR3 A0A0L7RAZ0 A0A1B6CXB3 A0A154PHN4 A0A158NVW0 D6WCR2 A0A0T6B1H0 A0A1B6CDG8 A0A0M8ZZM2 A0A1W4WBD0 A0A088A7Y0 A0A1B6E2R7 A0A2A3ENY0 A0A0J7KQX8 A0A291S6V9 A0A151ICI4 A0A195ESA0 E2BN97 A0A224XNL6 A0A0A9YM41 F4X4V6 A0A151HXY4 U5EZT8 A0A151X4W0 A0A1S3JGW5 A0A2J7RLY0 E2AX87 A0A336LNF8 N6TWY7 J9K6F6 A0A2H8TT50 A0A0L0CFN1 Q86MK0 A0A1I8MK24 A0A1B0CWH2 A0A0A1X767 T1HIF4 W8CBN0 A0A0K8UPS1 K1R5H7 A0A210QEX6 A0A084VM35 E0VGZ0 A0A034WG55 A0A182YNF0 M7C196 A0A0A0AJ35 A0A2G8K7L9 A0A182LWZ0 A0A182VUM5 A0A093F2H5 A0A1I8PRW0 K7FKI9 A0A1S4EQU6 A0A182IW47 A0A1Q3EVR6 A0A091QSK4 A0A3B0K649 A0A3M0K602 A0A1B0GCS7 A0A1A9UPX9 A0A091N3C8 A0A1S2ZXH4 A0A3L8SQ17 A0A091HQM6 A0A093C6Y9 A0A1A9WF02 A0A094KR47 A0A087V2G3 A0A093FZW3 A0A087RHH1 A0A2I0MC45 A0A1J1HSS0 E2RGS2 A0A091F6V4 A0A093NDJ8 A0A1L8GM26 A0A1L8GM25 A0A1U7T895 H0Z9X7 A0A1V4JX82 A0A091U3P8 A0A2C9JHF3 Q6P867 Q24044 G3TL12
A0A194RA82 A0A1B6GJ63 A0A1Y1LIH0 K7ISD3 A0A232F777 A0A026WIT0 A0A1B6CMN1 A0A1B6KWR3 A0A0L7RAZ0 A0A1B6CXB3 A0A154PHN4 A0A158NVW0 D6WCR2 A0A0T6B1H0 A0A1B6CDG8 A0A0M8ZZM2 A0A1W4WBD0 A0A088A7Y0 A0A1B6E2R7 A0A2A3ENY0 A0A0J7KQX8 A0A291S6V9 A0A151ICI4 A0A195ESA0 E2BN97 A0A224XNL6 A0A0A9YM41 F4X4V6 A0A151HXY4 U5EZT8 A0A151X4W0 A0A1S3JGW5 A0A2J7RLY0 E2AX87 A0A336LNF8 N6TWY7 J9K6F6 A0A2H8TT50 A0A0L0CFN1 Q86MK0 A0A1I8MK24 A0A1B0CWH2 A0A0A1X767 T1HIF4 W8CBN0 A0A0K8UPS1 K1R5H7 A0A210QEX6 A0A084VM35 E0VGZ0 A0A034WG55 A0A182YNF0 M7C196 A0A0A0AJ35 A0A2G8K7L9 A0A182LWZ0 A0A182VUM5 A0A093F2H5 A0A1I8PRW0 K7FKI9 A0A1S4EQU6 A0A182IW47 A0A1Q3EVR6 A0A091QSK4 A0A3B0K649 A0A3M0K602 A0A1B0GCS7 A0A1A9UPX9 A0A091N3C8 A0A1S2ZXH4 A0A3L8SQ17 A0A091HQM6 A0A093C6Y9 A0A1A9WF02 A0A094KR47 A0A087V2G3 A0A093FZW3 A0A087RHH1 A0A2I0MC45 A0A1J1HSS0 E2RGS2 A0A091F6V4 A0A093NDJ8 A0A1L8GM26 A0A1L8GM25 A0A1U7T895 H0Z9X7 A0A1V4JX82 A0A091U3P8 A0A2C9JHF3 Q6P867 Q24044 G3TL12
Pubmed
19121390
22118469
23622113
26354079
28004739
20075255
+ More
28648823 24508170 21347285 18362917 19820115 28992199 20798317 25401762 26823975 21719571 23537049 26108605 25315136 25830018 24495485 22992520 28812685 24438588 20566863 25348373 25244985 23624526 29023486 17381049 30282656 23371554 16341006 27762356 20360741 15562597 7730407 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
28648823 24508170 21347285 18362917 19820115 28992199 20798317 25401762 26823975 21719571 23537049 26108605 25315136 25830018 24495485 22992520 28812685 24438588 20566863 25348373 25244985 23624526 29023486 17381049 30282656 23371554 16341006 27762356 20360741 15562597 7730407 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
NWSH01000423
PCG76499.1
ODYU01004759
SOQ44983.1
RSAL01000076
RVE48845.1
+ More
BABH01019457 BABH01019458 BABH01019459 AGBW02012218 OWR45426.1 GAIX01003506 JAA89054.1 KQ460473 KPJ14414.1 GECZ01007291 JAS62478.1 GEZM01059724 JAV71386.1 NNAY01000827 OXU26298.1 KK107231 EZA55009.1 GEDC01022598 GEDC01014258 JAS14700.1 JAS23040.1 GEBQ01024099 JAT15878.1 KQ414617 KOC68087.1 GEDC01019211 GEDC01001610 JAS18087.1 JAS35688.1 KQ434899 KZC11004.1 ADTU01027543 KQ971317 EEZ98810.1 LJIG01016407 KRT80695.1 GEDC01025802 JAS11496.1 KQ435793 KOX74167.1 GEDC01005075 JAS32223.1 KZ288203 PBC33427.1 LBMM01004153 KMQ92753.1 MF432992 ATL75343.1 KQ978069 KYM97362.1 KQ981993 KYN31103.1 GL449385 EFN82859.1 GFTR01006847 JAW09579.1 GBHO01010320 GBRD01017692 GDHC01020438 JAG33284.1 JAG48135.1 JAP98190.1 GL888679 EGI58529.1 KQ976730 KYM76421.1 GANO01000168 JAB59703.1 KQ982522 KYQ55456.1 NEVH01002587 PNF41841.1 GL443520 EFN61970.1 UFQT01000063 SSX19285.1 APGK01048500 KB741112 KB632184 ENN73820.1 ERL89629.1 ABLF02011612 GFXV01005335 MBW17140.1 JRES01000539 KNC30299.1 AY236483 AAO85336.1 AJWK01032468 GBXI01007571 JAD06721.1 ACPB03001249 GAMC01000369 JAC06187.1 GDHF01023620 JAI28694.1 JH816764 EKC38794.1 NEDP02003955 OWF47285.1 ATLV01014577 KE524975 KFB39029.1 DS235154 EEB12646.1 GAKP01005283 JAC53669.1 KB533688 EMP34162.1 KL871737 KGL93035.1 MRZV01000808 PIK44016.1 AXCM01012412 KK387070 KFV51792.1 AGCU01029742 AGCU01029743 GFDL01015652 JAV19393.1 KK705742 KFQ30168.1 OUUW01000006 SPP81479.1 QRBI01000117 RMC08568.1 CCAG010002220 KL376153 KFP83946.1 QUSF01000010 RLW05850.1 KL217788 KFO98588.1 KL240502 KFV11700.1 KL352511 KFZ59832.1 KL477420 KFO06805.1 KK638743 KFV59914.1 KL226373 KFM12925.1 AKCR02000021 PKK27251.1 CVRI01000020 CRK91120.1 AAEX03009812 AAEX03009813 KK719659 KFO64567.1 KL224623 KFW62241.1 CM004472 OCT84890.1 OCT84889.1 ABQF01041516 LSYS01005497 OPJ76789.1 OPJ76790.1 KK497664 KFQ66449.1 BC061363 CR760268 AAH61363.1 CAJ83291.1 U22419 AE014134 BT029432 BT056243 AAA83150.1 AAF53523.1 ABK57089.1 ACL68690.1
BABH01019457 BABH01019458 BABH01019459 AGBW02012218 OWR45426.1 GAIX01003506 JAA89054.1 KQ460473 KPJ14414.1 GECZ01007291 JAS62478.1 GEZM01059724 JAV71386.1 NNAY01000827 OXU26298.1 KK107231 EZA55009.1 GEDC01022598 GEDC01014258 JAS14700.1 JAS23040.1 GEBQ01024099 JAT15878.1 KQ414617 KOC68087.1 GEDC01019211 GEDC01001610 JAS18087.1 JAS35688.1 KQ434899 KZC11004.1 ADTU01027543 KQ971317 EEZ98810.1 LJIG01016407 KRT80695.1 GEDC01025802 JAS11496.1 KQ435793 KOX74167.1 GEDC01005075 JAS32223.1 KZ288203 PBC33427.1 LBMM01004153 KMQ92753.1 MF432992 ATL75343.1 KQ978069 KYM97362.1 KQ981993 KYN31103.1 GL449385 EFN82859.1 GFTR01006847 JAW09579.1 GBHO01010320 GBRD01017692 GDHC01020438 JAG33284.1 JAG48135.1 JAP98190.1 GL888679 EGI58529.1 KQ976730 KYM76421.1 GANO01000168 JAB59703.1 KQ982522 KYQ55456.1 NEVH01002587 PNF41841.1 GL443520 EFN61970.1 UFQT01000063 SSX19285.1 APGK01048500 KB741112 KB632184 ENN73820.1 ERL89629.1 ABLF02011612 GFXV01005335 MBW17140.1 JRES01000539 KNC30299.1 AY236483 AAO85336.1 AJWK01032468 GBXI01007571 JAD06721.1 ACPB03001249 GAMC01000369 JAC06187.1 GDHF01023620 JAI28694.1 JH816764 EKC38794.1 NEDP02003955 OWF47285.1 ATLV01014577 KE524975 KFB39029.1 DS235154 EEB12646.1 GAKP01005283 JAC53669.1 KB533688 EMP34162.1 KL871737 KGL93035.1 MRZV01000808 PIK44016.1 AXCM01012412 KK387070 KFV51792.1 AGCU01029742 AGCU01029743 GFDL01015652 JAV19393.1 KK705742 KFQ30168.1 OUUW01000006 SPP81479.1 QRBI01000117 RMC08568.1 CCAG010002220 KL376153 KFP83946.1 QUSF01000010 RLW05850.1 KL217788 KFO98588.1 KL240502 KFV11700.1 KL352511 KFZ59832.1 KL477420 KFO06805.1 KK638743 KFV59914.1 KL226373 KFM12925.1 AKCR02000021 PKK27251.1 CVRI01000020 CRK91120.1 AAEX03009812 AAEX03009813 KK719659 KFO64567.1 KL224623 KFW62241.1 CM004472 OCT84890.1 OCT84889.1 ABQF01041516 LSYS01005497 OPJ76789.1 OPJ76790.1 KK497664 KFQ66449.1 BC061363 CR760268 AAH61363.1 CAJ83291.1 U22419 AE014134 BT029432 BT056243 AAA83150.1 AAF53523.1 ABK57089.1 ACL68690.1
Proteomes
UP000218220
UP000283053
UP000005204
UP000007151
UP000053240
UP000002358
+ More
UP000215335 UP000053097 UP000053825 UP000076502 UP000005205 UP000007266 UP000053105 UP000192223 UP000005203 UP000242457 UP000036403 UP000078542 UP000078541 UP000008237 UP000007755 UP000078540 UP000075809 UP000085678 UP000235965 UP000000311 UP000019118 UP000030742 UP000007819 UP000037069 UP000095301 UP000092461 UP000015103 UP000005408 UP000242188 UP000030765 UP000009046 UP000076408 UP000031443 UP000053858 UP000230750 UP000075883 UP000075920 UP000095300 UP000007267 UP000079169 UP000075880 UP000268350 UP000269221 UP000092444 UP000078200 UP000079721 UP000276834 UP000054308 UP000091820 UP000053286 UP000053872 UP000183832 UP000002254 UP000052976 UP000054081 UP000186698 UP000189704 UP000007754 UP000190648 UP000076420 UP000000803 UP000007646
UP000215335 UP000053097 UP000053825 UP000076502 UP000005205 UP000007266 UP000053105 UP000192223 UP000005203 UP000242457 UP000036403 UP000078542 UP000078541 UP000008237 UP000007755 UP000078540 UP000075809 UP000085678 UP000235965 UP000000311 UP000019118 UP000030742 UP000007819 UP000037069 UP000095301 UP000092461 UP000015103 UP000005408 UP000242188 UP000030765 UP000009046 UP000076408 UP000031443 UP000053858 UP000230750 UP000075883 UP000075920 UP000095300 UP000007267 UP000079169 UP000075880 UP000268350 UP000269221 UP000092444 UP000078200 UP000079721 UP000276834 UP000054308 UP000091820 UP000053286 UP000053872 UP000183832 UP000002254 UP000052976 UP000054081 UP000186698 UP000189704 UP000007754 UP000190648 UP000076420 UP000000803 UP000007646
Pfam
Interpro
IPR015943
WD40/YVTN_repeat-like_dom_sf
+ More
IPR001680 WD40_repeat
IPR033010 Cdc20/Fizzy
IPR017986 WD40_repeat_dom
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR011029 DEATH-like_dom_sf
IPR001315 CARD
IPR036388 WH-like_DNA-bd_sf
IPR041452 APAF1_C
IPR011047 Quinoprotein_ADH-like_supfam
IPR027417 P-loop_NTPase
IPR002182 NB-ARC
IPR001900 RNase_II/R
IPR041093 Dis3l2_C
IPR041505 Dis3_CSD2
IPR022966 RNase_II/R_CS
IPR012340 NA-bd_OB-fold
IPR024977 Apc4_WD40_dom
IPR001680 WD40_repeat
IPR033010 Cdc20/Fizzy
IPR017986 WD40_repeat_dom
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR011029 DEATH-like_dom_sf
IPR001315 CARD
IPR036388 WH-like_DNA-bd_sf
IPR041452 APAF1_C
IPR011047 Quinoprotein_ADH-like_supfam
IPR027417 P-loop_NTPase
IPR002182 NB-ARC
IPR001900 RNase_II/R
IPR041093 Dis3l2_C
IPR041505 Dis3_CSD2
IPR022966 RNase_II/R_CS
IPR012340 NA-bd_OB-fold
IPR024977 Apc4_WD40_dom
SUPFAM
Gene 3D
ProteinModelPortal
A0A2A4JXH4
A0A2H1VW01
A0A3S2NZY5
H9JNC5
A0A212EVB1
S4PDU2
+ More
A0A194RA82 A0A1B6GJ63 A0A1Y1LIH0 K7ISD3 A0A232F777 A0A026WIT0 A0A1B6CMN1 A0A1B6KWR3 A0A0L7RAZ0 A0A1B6CXB3 A0A154PHN4 A0A158NVW0 D6WCR2 A0A0T6B1H0 A0A1B6CDG8 A0A0M8ZZM2 A0A1W4WBD0 A0A088A7Y0 A0A1B6E2R7 A0A2A3ENY0 A0A0J7KQX8 A0A291S6V9 A0A151ICI4 A0A195ESA0 E2BN97 A0A224XNL6 A0A0A9YM41 F4X4V6 A0A151HXY4 U5EZT8 A0A151X4W0 A0A1S3JGW5 A0A2J7RLY0 E2AX87 A0A336LNF8 N6TWY7 J9K6F6 A0A2H8TT50 A0A0L0CFN1 Q86MK0 A0A1I8MK24 A0A1B0CWH2 A0A0A1X767 T1HIF4 W8CBN0 A0A0K8UPS1 K1R5H7 A0A210QEX6 A0A084VM35 E0VGZ0 A0A034WG55 A0A182YNF0 M7C196 A0A0A0AJ35 A0A2G8K7L9 A0A182LWZ0 A0A182VUM5 A0A093F2H5 A0A1I8PRW0 K7FKI9 A0A1S4EQU6 A0A182IW47 A0A1Q3EVR6 A0A091QSK4 A0A3B0K649 A0A3M0K602 A0A1B0GCS7 A0A1A9UPX9 A0A091N3C8 A0A1S2ZXH4 A0A3L8SQ17 A0A091HQM6 A0A093C6Y9 A0A1A9WF02 A0A094KR47 A0A087V2G3 A0A093FZW3 A0A087RHH1 A0A2I0MC45 A0A1J1HSS0 E2RGS2 A0A091F6V4 A0A093NDJ8 A0A1L8GM26 A0A1L8GM25 A0A1U7T895 H0Z9X7 A0A1V4JX82 A0A091U3P8 A0A2C9JHF3 Q6P867 Q24044 G3TL12
A0A194RA82 A0A1B6GJ63 A0A1Y1LIH0 K7ISD3 A0A232F777 A0A026WIT0 A0A1B6CMN1 A0A1B6KWR3 A0A0L7RAZ0 A0A1B6CXB3 A0A154PHN4 A0A158NVW0 D6WCR2 A0A0T6B1H0 A0A1B6CDG8 A0A0M8ZZM2 A0A1W4WBD0 A0A088A7Y0 A0A1B6E2R7 A0A2A3ENY0 A0A0J7KQX8 A0A291S6V9 A0A151ICI4 A0A195ESA0 E2BN97 A0A224XNL6 A0A0A9YM41 F4X4V6 A0A151HXY4 U5EZT8 A0A151X4W0 A0A1S3JGW5 A0A2J7RLY0 E2AX87 A0A336LNF8 N6TWY7 J9K6F6 A0A2H8TT50 A0A0L0CFN1 Q86MK0 A0A1I8MK24 A0A1B0CWH2 A0A0A1X767 T1HIF4 W8CBN0 A0A0K8UPS1 K1R5H7 A0A210QEX6 A0A084VM35 E0VGZ0 A0A034WG55 A0A182YNF0 M7C196 A0A0A0AJ35 A0A2G8K7L9 A0A182LWZ0 A0A182VUM5 A0A093F2H5 A0A1I8PRW0 K7FKI9 A0A1S4EQU6 A0A182IW47 A0A1Q3EVR6 A0A091QSK4 A0A3B0K649 A0A3M0K602 A0A1B0GCS7 A0A1A9UPX9 A0A091N3C8 A0A1S2ZXH4 A0A3L8SQ17 A0A091HQM6 A0A093C6Y9 A0A1A9WF02 A0A094KR47 A0A087V2G3 A0A093FZW3 A0A087RHH1 A0A2I0MC45 A0A1J1HSS0 E2RGS2 A0A091F6V4 A0A093NDJ8 A0A1L8GM26 A0A1L8GM25 A0A1U7T895 H0Z9X7 A0A1V4JX82 A0A091U3P8 A0A2C9JHF3 Q6P867 Q24044 G3TL12
PDB
6F0X
E-value=5.00656e-137,
Score=1251
Ontologies
GO
GO:0010997
GO:1904668
GO:0031145
GO:0097027
GO:0043531
GO:0042981
GO:0051301
GO:0004540
GO:0003723
GO:0003743
GO:0004402
GO:0019899
GO:0008022
GO:0005654
GO:0005829
GO:0005680
GO:0090307
GO:0007064
GO:0031915
GO:0040020
GO:0090129
GO:0007147
GO:0000278
GO:0060547
GO:0000776
GO:0005813
GO:0061630
GO:0030718
GO:0007144
GO:0007096
GO:0097150
GO:0005819
GO:0005737
GO:0030163
GO:0030162
GO:0005515
GO:0006836
GO:0016021
GO:0051276
GO:0016491
GO:0016620
GO:0016787
GO:0003676
GO:0015930
PANTHER
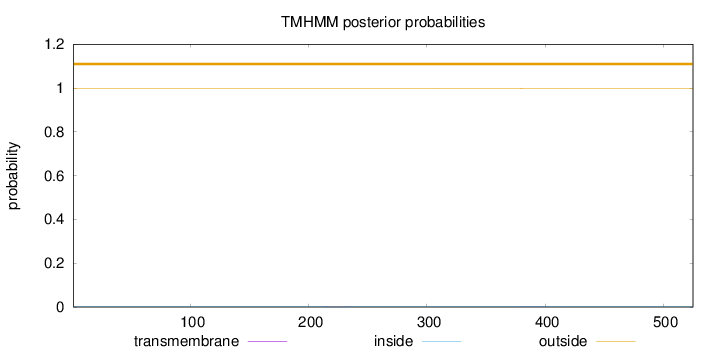
Topology
Length:
525
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0344
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00076
outside
1 - 525
Population Genetic Test Statistics
Pi
247.055069
Theta
189.367262
Tajima's D
0.832053
CLR
14.679702
CSRT
0.615469226538673
Interpretation
Uncertain
