Gene
KWMTBOMO13668 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011029
Annotation
PREDICTED:_acetyl-CoA_acetyltransferase?_mitochondrial_[Papilio_polytes]
Location in the cell
Cytoplasmic Reliability : 1.23 Mitochondrial Reliability : 1.529
Sequence
CDS
ATGAAGCCAATATTTACAGCAATGGCAGCATTCTCGACCAAAGTTTCCCTCAACGAAGTAGTAATTGCATCAGCAGTTAGAACTCCTATGGGTTCTTTCCGAGGGAGCTTATCTAGTTTATCAGCATCAGAATTAGGCGCTGTCGCTGTTAACGCGGCAATCGAAAGAGCCGGTATTCCTAAAGAAGAAATAAAAGAGGTCTACATTGGCAATGTTTGTTCTGCAAATTTGGGCCAAGCACCTGCAAGACAAGCTGTAATATTTGCAGGTTTGCCAAAAAGTACCATATGTACAACTGTAAACAAAGTATGTGCCTCTGGCATGAAATCTATAATGTTGGCAGCACAAGGTCTACAAACTGGAGCCCAAGATATAATACTTGCTGGTGGGATGGAATCTATGTCAAATGTACCTTTTTATTTGAAGAGAGGTGAAACTTCATATGGAGGAATGCAGTTAGTTGATGGAATAGTGTTTGATGGGCTCACAGATGTGTACAACAAATTTCACATGGGAAATTGTGCTGAAAACACAGCAAAAAAATTACAAATTACTAGACAAGATCAAGATGAATATGCTGTCAATAGTTACAAGAGAAGTGCAGCTGCTTATGAAGCTAAAGCTTTTGTTGATGAACTTGTACCAGTACCCGTTCCACAGAAAAGAGGGGCTCCTGTAATATTTGCTGAAGATGAAGAGTATAAAAGAGTTAATTTTGAAAAGTTTACAAAATTGTCTACGGTATTCCAAAAGGAAAATGGCACTGTTACTGCAGGCAATGCTTCAACATTAAATGATGGAGCTGCAGCTTTAGTGTTAATGACGGCTGATGCCGCTAAAAGATTAAATGTCAAGCCGATTGCCCGAATTGTTGGATTTGCTGATGGGGAATGTGATCCCATTGACTTCCCAATAGCACCAGCGGTTGCCATTCCTAAATTATTAGAGAAGACTGGTGTCAGGAAGGAAGATGTTGCATTGTGGGAGATAAATGAAGCTTTCAGTGTTGTTGCTGTTGCAAATCAAAAATTATTAGGATTAGATCCATCAAAAATTAATGTACATGGTGGAGCTGTCAGTTTGGGTCATCCAATTGGTATGTCTGGAGCTCGCATTGTTGTACATCTGTGCCATGCTTTAAAAAAAGGAGAAAAGGGAGTTGCTTCAATTTGTAATGGTGGAGGTGGTGCATCATCTGTAATGATTGAAAAAATAGCCGAAACTCCAGAAGAGCTACCGGTTTTGACATTTTATACCAAAGATCCATGTGGTCTCTGTGATTTAGTTATGGAAGACCTTGAACCATATAAAAAGAGACTTATCATACAAAAAATTGATATCACTCAAAAAGAAAATTTGCGGTGGTTACGCCTTTATAGACTTGAAATACCAGTCTTATTCTTGAATGGACATTTTTTATGTAAACATAAGTTAAACAAAGATCTTCTTGAGAAAAAGTTGACATTAATTGAAATAGAAAAAAAACAAACATCATAA
Protein
MKPIFTAMAAFSTKVSLNEVVIASAVRTPMGSFRGSLSSLSASELGAVAVNAAIERAGIPKEEIKEVYIGNVCSANLGQAPARQAVIFAGLPKSTICTTVNKVCASGMKSIMLAAQGLQTGAQDIILAGGMESMSNVPFYLKRGETSYGGMQLVDGIVFDGLTDVYNKFHMGNCAENTAKKLQITRQDQDEYAVNSYKRSAAAYEAKAFVDELVPVPVPQKRGAPVIFAEDEEYKRVNFEKFTKLSTVFQKENGTVTAGNASTLNDGAAALVLMTADAAKRLNVKPIARIVGFADGECDPIDFPIAPAVAIPKLLEKTGVRKEDVALWEINEAFSVVAVANQKLLGLDPSKINVHGGAVSLGHPIGMSGARIVVHLCHALKKGEKGVASICNGGGGASSVMIEKIAETPEELPVLTFYTKDPCGLCDLVMEDLEPYKKRLIIQKIDITQKENLRWLRLYRLEIPVLFLNGHFLCKHKLNKDLLEKKLTLIEIEKKQTS
Summary
Similarity
Belongs to the thiolase-like superfamily. Thiolase family.
Uniprot
A0A194PSL1
A0A212EV98
A0A194R9U5
A0A2H1V442
A0A088M985
A0A0L7K3X0
+ More
A0A1Q3G5I6 H9JNC6 A5A797 D6WAN7 U4UXZ8 S4PX78 A0A026WB42 A0A0L7RB84 E2A7A4 A0A232F866 I4DNZ9 A0A195CZR7 A0A1V0M897 A0A151X3E1 A0A084WCJ7 A0A195ERW0 A0A182Y6Z9 A0A182J461 E2BF82 A0A195BR39 W5JHX2 A0A154PGM7 A0A182H3Q8 A0A182QXB2 A0A182SRZ0 A0A182FTX4 A0A2A4JWV9 A0A182N3D0 A0A182RLJ8 A0A182ML35 A0A182W1L8 A0A291P161 A0A291P121 A0A2H4UZK2 A0A088A7V7 A0A182U9K0 A0A1B6J2A8 A0A182HY56 A0A182WXI1 A0A182UML4 A0A182LHA8 B0XJ38 A0A182P2N2 A0A195EF02 A0A067QU33 A0A1B6D1S8 A0A336LLV7 A0A2A3EPK3 A0A1J1J5R4 A0A1L8DYX1 A0A1L8DZ47 K7ISJ1 A0A023ETB1 I1VJ17 A0A1L8DZ78 A0A1L8DYQ0 F4WQU2 A0A158NRR5 A0A1W4XE81 A0A0H4IRH6 A0A0M8ZZA5 A0A0C9QQV1 A0A1Y1M1H4 Q56CY6 A0A2M3ZH75 A0A2M3ZH66 A0A2H1V0U2 A0A1V0M8A0 A0A0P6H5P3 A0A2M4ADC3 A0A2M4BPR8 A0A0P5IFM7 A0A0P6D8S6 A0A2M4ADB4 Q175J7 A0A1B6GLL4 A0A0P5QQV0 A0A0P5XC51 A0A1S4FE42 T1E9X1 A0A2M4AD81 A0A0P5IJ91 A0A1B6FXX3 A0A0P0DZ45 A0A0L0BRE9 A0A1B6E2E7 A0A1B6J834 A0A1A9X5A9 A0A2M4BPJ3 A0A0P5GAD7 A0A0P5X6I4 A0A0P5GQD5 U5ETW3 T1PDZ7
A0A1Q3G5I6 H9JNC6 A5A797 D6WAN7 U4UXZ8 S4PX78 A0A026WB42 A0A0L7RB84 E2A7A4 A0A232F866 I4DNZ9 A0A195CZR7 A0A1V0M897 A0A151X3E1 A0A084WCJ7 A0A195ERW0 A0A182Y6Z9 A0A182J461 E2BF82 A0A195BR39 W5JHX2 A0A154PGM7 A0A182H3Q8 A0A182QXB2 A0A182SRZ0 A0A182FTX4 A0A2A4JWV9 A0A182N3D0 A0A182RLJ8 A0A182ML35 A0A182W1L8 A0A291P161 A0A291P121 A0A2H4UZK2 A0A088A7V7 A0A182U9K0 A0A1B6J2A8 A0A182HY56 A0A182WXI1 A0A182UML4 A0A182LHA8 B0XJ38 A0A182P2N2 A0A195EF02 A0A067QU33 A0A1B6D1S8 A0A336LLV7 A0A2A3EPK3 A0A1J1J5R4 A0A1L8DYX1 A0A1L8DZ47 K7ISJ1 A0A023ETB1 I1VJ17 A0A1L8DZ78 A0A1L8DYQ0 F4WQU2 A0A158NRR5 A0A1W4XE81 A0A0H4IRH6 A0A0M8ZZA5 A0A0C9QQV1 A0A1Y1M1H4 Q56CY6 A0A2M3ZH75 A0A2M3ZH66 A0A2H1V0U2 A0A1V0M8A0 A0A0P6H5P3 A0A2M4ADC3 A0A2M4BPR8 A0A0P5IFM7 A0A0P6D8S6 A0A2M4ADB4 Q175J7 A0A1B6GLL4 A0A0P5QQV0 A0A0P5XC51 A0A1S4FE42 T1E9X1 A0A2M4AD81 A0A0P5IJ91 A0A1B6FXX3 A0A0P0DZ45 A0A0L0BRE9 A0A1B6E2E7 A0A1B6J834 A0A1A9X5A9 A0A2M4BPJ3 A0A0P5GAD7 A0A0P5X6I4 A0A0P5GQD5 U5ETW3 T1PDZ7
Pubmed
26354079
22118469
26385554
26227816
19121390
17628279
+ More
18362917 19820115 23537049 23622113 24508170 30249741 20798317 28648823 22651552 28349297 24438588 25244985 20920257 23761445 26483478 28986331 29185447 20966253 24845553 20075255 24945155 22516182 21719571 21347285 26899871 28004739 17510324 26445454 26108605 25315136
18362917 19820115 23537049 23622113 24508170 30249741 20798317 28648823 22651552 28349297 24438588 25244985 20920257 23761445 26483478 28986331 29185447 20966253 24845553 20075255 24945155 22516182 21719571 21347285 26899871 28004739 17510324 26445454 26108605 25315136
EMBL
KQ459600
KPI94110.1
AGBW02012218
OWR45428.1
KQ460473
KPJ14412.1
+ More
ODYU01000581 SOQ35615.1 KJ579206 AIN34682.1 JTDY01011334 KOB56897.1 GEYN01000174 JAV44815.1 BABH01019456 AB274988 BAF62106.1 KQ971312 EEZ97970.1 KB632410 ERL95190.1 GAIX01005513 JAA87047.1 KK107347 QOIP01000007 EZA52249.1 RLU20939.1 KQ414617 KOC68080.1 GL437329 EFN70601.1 NNAY01000759 OXU26649.1 AK403218 BAM19639.1 KQ977041 KYN06116.1 KU755506 ARD71216.1 KQ982562 KYQ54912.1 ATLV01022681 KE525335 KFB47941.1 KQ981993 KYN30916.1 GL447952 EFN85668.1 KQ976424 KYM88603.1 ADMH02001457 ETN62505.1 KQ434899 KZC11011.1 JXUM01107962 JXUM01107963 JXUM01107964 JXUM01107965 JXUM01107966 JXUM01107967 JXUM01107968 KQ565188 KXJ71243.1 AXCN02001111 NWSH01000423 PCG76497.1 AXCM01006132 MF706171 ATJ44598.1 MF687639 ATJ44565.1 KY971766 AUA18033.1 GECU01014391 JAS93315.1 APCN01004493 DS233428 EDS29933.1 KQ978983 KYN26818.1 KK853270 KDR09162.1 GEDC01017649 JAS19649.1 UFQS01000042 UFQT01000042 SSW98319.1 SSX18705.1 KZ288203 PBC33434.1 CVRI01000073 CRL07747.1 GFDF01002414 JAV11670.1 GFDF01002385 JAV11699.1 GAPW01001060 JAC12538.1 JQ855638 AFI45001.1 GFDF01002492 JAV11592.1 GFDF01002491 JAV11593.1 GL888275 EGI63455.1 ADTU01024274 KP689334 AKO63316.1 KQ435793 KOX74160.1 GBYB01006109 JAG75876.1 GEZM01042666 JAV79742.1 AY966010 AAX78436.1 GGFM01007037 MBW27788.1 GGFM01007027 MBW27778.1 ODYU01000159 SOQ34458.1 KU755503 ARD71213.1 GDIQ01036854 JAN57883.1 GGFK01005401 MBW38722.1 GGFJ01005861 MBW55002.1 GDIQ01214056 GDIQ01109284 JAK37669.1 GDIQ01081802 JAN12935.1 GGFK01005391 MBW38712.1 CH477399 EAT41757.1 GECZ01006479 JAS63290.1 GDIQ01131134 JAL20592.1 GDIP01074264 LRGB01000115 JAM29451.1 KZS20723.1 GAMD01001528 JAB00063.1 GGFK01005425 MBW38746.1 GDIQ01214057 JAK37668.1 GECZ01014721 GECZ01003669 JAS55048.1 JAS66100.1 KT261712 ALJ30252.1 JRES01001480 KNC22563.1 GEDC01005219 JAS32079.1 GECU01034670 GECU01018604 GECU01012360 JAS73036.1 JAS89102.1 JAS95346.1 GGFJ01005864 MBW55005.1 GDIQ01247346 JAK04379.1 GDIP01088713 JAM15002.1 GDIQ01241102 JAK10623.1 GANO01002599 JAB57272.1 KA646977 AFP61606.1
ODYU01000581 SOQ35615.1 KJ579206 AIN34682.1 JTDY01011334 KOB56897.1 GEYN01000174 JAV44815.1 BABH01019456 AB274988 BAF62106.1 KQ971312 EEZ97970.1 KB632410 ERL95190.1 GAIX01005513 JAA87047.1 KK107347 QOIP01000007 EZA52249.1 RLU20939.1 KQ414617 KOC68080.1 GL437329 EFN70601.1 NNAY01000759 OXU26649.1 AK403218 BAM19639.1 KQ977041 KYN06116.1 KU755506 ARD71216.1 KQ982562 KYQ54912.1 ATLV01022681 KE525335 KFB47941.1 KQ981993 KYN30916.1 GL447952 EFN85668.1 KQ976424 KYM88603.1 ADMH02001457 ETN62505.1 KQ434899 KZC11011.1 JXUM01107962 JXUM01107963 JXUM01107964 JXUM01107965 JXUM01107966 JXUM01107967 JXUM01107968 KQ565188 KXJ71243.1 AXCN02001111 NWSH01000423 PCG76497.1 AXCM01006132 MF706171 ATJ44598.1 MF687639 ATJ44565.1 KY971766 AUA18033.1 GECU01014391 JAS93315.1 APCN01004493 DS233428 EDS29933.1 KQ978983 KYN26818.1 KK853270 KDR09162.1 GEDC01017649 JAS19649.1 UFQS01000042 UFQT01000042 SSW98319.1 SSX18705.1 KZ288203 PBC33434.1 CVRI01000073 CRL07747.1 GFDF01002414 JAV11670.1 GFDF01002385 JAV11699.1 GAPW01001060 JAC12538.1 JQ855638 AFI45001.1 GFDF01002492 JAV11592.1 GFDF01002491 JAV11593.1 GL888275 EGI63455.1 ADTU01024274 KP689334 AKO63316.1 KQ435793 KOX74160.1 GBYB01006109 JAG75876.1 GEZM01042666 JAV79742.1 AY966010 AAX78436.1 GGFM01007037 MBW27788.1 GGFM01007027 MBW27778.1 ODYU01000159 SOQ34458.1 KU755503 ARD71213.1 GDIQ01036854 JAN57883.1 GGFK01005401 MBW38722.1 GGFJ01005861 MBW55002.1 GDIQ01214056 GDIQ01109284 JAK37669.1 GDIQ01081802 JAN12935.1 GGFK01005391 MBW38712.1 CH477399 EAT41757.1 GECZ01006479 JAS63290.1 GDIQ01131134 JAL20592.1 GDIP01074264 LRGB01000115 JAM29451.1 KZS20723.1 GAMD01001528 JAB00063.1 GGFK01005425 MBW38746.1 GDIQ01214057 JAK37668.1 GECZ01014721 GECZ01003669 JAS55048.1 JAS66100.1 KT261712 ALJ30252.1 JRES01001480 KNC22563.1 GEDC01005219 JAS32079.1 GECU01034670 GECU01018604 GECU01012360 JAS73036.1 JAS89102.1 JAS95346.1 GGFJ01005864 MBW55005.1 GDIQ01247346 JAK04379.1 GDIP01088713 JAM15002.1 GDIQ01241102 JAK10623.1 GANO01002599 JAB57272.1 KA646977 AFP61606.1
Proteomes
UP000053268
UP000007151
UP000053240
UP000037510
UP000005204
UP000007266
+ More
UP000030742 UP000053097 UP000279307 UP000053825 UP000000311 UP000215335 UP000078542 UP000075809 UP000030765 UP000078541 UP000076408 UP000075880 UP000008237 UP000078540 UP000000673 UP000076502 UP000069940 UP000249989 UP000075886 UP000075901 UP000069272 UP000218220 UP000075884 UP000075900 UP000075883 UP000075920 UP000005203 UP000075902 UP000075840 UP000076407 UP000075903 UP000075882 UP000002320 UP000075885 UP000078492 UP000027135 UP000242457 UP000183832 UP000002358 UP000007755 UP000005205 UP000192223 UP000053105 UP000008820 UP000076858 UP000037069 UP000091820 UP000095301
UP000030742 UP000053097 UP000279307 UP000053825 UP000000311 UP000215335 UP000078542 UP000075809 UP000030765 UP000078541 UP000076408 UP000075880 UP000008237 UP000078540 UP000000673 UP000076502 UP000069940 UP000249989 UP000075886 UP000075901 UP000069272 UP000218220 UP000075884 UP000075900 UP000075883 UP000075920 UP000005203 UP000075902 UP000075840 UP000076407 UP000075903 UP000075882 UP000002320 UP000075885 UP000078492 UP000027135 UP000242457 UP000183832 UP000002358 UP000007755 UP000005205 UP000192223 UP000053105 UP000008820 UP000076858 UP000037069 UP000091820 UP000095301
Interpro
Gene 3D
CDD
ProteinModelPortal
A0A194PSL1
A0A212EV98
A0A194R9U5
A0A2H1V442
A0A088M985
A0A0L7K3X0
+ More
A0A1Q3G5I6 H9JNC6 A5A797 D6WAN7 U4UXZ8 S4PX78 A0A026WB42 A0A0L7RB84 E2A7A4 A0A232F866 I4DNZ9 A0A195CZR7 A0A1V0M897 A0A151X3E1 A0A084WCJ7 A0A195ERW0 A0A182Y6Z9 A0A182J461 E2BF82 A0A195BR39 W5JHX2 A0A154PGM7 A0A182H3Q8 A0A182QXB2 A0A182SRZ0 A0A182FTX4 A0A2A4JWV9 A0A182N3D0 A0A182RLJ8 A0A182ML35 A0A182W1L8 A0A291P161 A0A291P121 A0A2H4UZK2 A0A088A7V7 A0A182U9K0 A0A1B6J2A8 A0A182HY56 A0A182WXI1 A0A182UML4 A0A182LHA8 B0XJ38 A0A182P2N2 A0A195EF02 A0A067QU33 A0A1B6D1S8 A0A336LLV7 A0A2A3EPK3 A0A1J1J5R4 A0A1L8DYX1 A0A1L8DZ47 K7ISJ1 A0A023ETB1 I1VJ17 A0A1L8DZ78 A0A1L8DYQ0 F4WQU2 A0A158NRR5 A0A1W4XE81 A0A0H4IRH6 A0A0M8ZZA5 A0A0C9QQV1 A0A1Y1M1H4 Q56CY6 A0A2M3ZH75 A0A2M3ZH66 A0A2H1V0U2 A0A1V0M8A0 A0A0P6H5P3 A0A2M4ADC3 A0A2M4BPR8 A0A0P5IFM7 A0A0P6D8S6 A0A2M4ADB4 Q175J7 A0A1B6GLL4 A0A0P5QQV0 A0A0P5XC51 A0A1S4FE42 T1E9X1 A0A2M4AD81 A0A0P5IJ91 A0A1B6FXX3 A0A0P0DZ45 A0A0L0BRE9 A0A1B6E2E7 A0A1B6J834 A0A1A9X5A9 A0A2M4BPJ3 A0A0P5GAD7 A0A0P5X6I4 A0A0P5GQD5 U5ETW3 T1PDZ7
A0A1Q3G5I6 H9JNC6 A5A797 D6WAN7 U4UXZ8 S4PX78 A0A026WB42 A0A0L7RB84 E2A7A4 A0A232F866 I4DNZ9 A0A195CZR7 A0A1V0M897 A0A151X3E1 A0A084WCJ7 A0A195ERW0 A0A182Y6Z9 A0A182J461 E2BF82 A0A195BR39 W5JHX2 A0A154PGM7 A0A182H3Q8 A0A182QXB2 A0A182SRZ0 A0A182FTX4 A0A2A4JWV9 A0A182N3D0 A0A182RLJ8 A0A182ML35 A0A182W1L8 A0A291P161 A0A291P121 A0A2H4UZK2 A0A088A7V7 A0A182U9K0 A0A1B6J2A8 A0A182HY56 A0A182WXI1 A0A182UML4 A0A182LHA8 B0XJ38 A0A182P2N2 A0A195EF02 A0A067QU33 A0A1B6D1S8 A0A336LLV7 A0A2A3EPK3 A0A1J1J5R4 A0A1L8DYX1 A0A1L8DZ47 K7ISJ1 A0A023ETB1 I1VJ17 A0A1L8DZ78 A0A1L8DYQ0 F4WQU2 A0A158NRR5 A0A1W4XE81 A0A0H4IRH6 A0A0M8ZZA5 A0A0C9QQV1 A0A1Y1M1H4 Q56CY6 A0A2M3ZH75 A0A2M3ZH66 A0A2H1V0U2 A0A1V0M8A0 A0A0P6H5P3 A0A2M4ADC3 A0A2M4BPR8 A0A0P5IFM7 A0A0P6D8S6 A0A2M4ADB4 Q175J7 A0A1B6GLL4 A0A0P5QQV0 A0A0P5XC51 A0A1S4FE42 T1E9X1 A0A2M4AD81 A0A0P5IJ91 A0A1B6FXX3 A0A0P0DZ45 A0A0L0BRE9 A0A1B6E2E7 A0A1B6J834 A0A1A9X5A9 A0A2M4BPJ3 A0A0P5GAD7 A0A0P5X6I4 A0A0P5GQD5 U5ETW3 T1PDZ7
PDB
2IBY
E-value=5.6798e-135,
Score=1233
Ontologies
PATHWAY
00071
Fatty acid degradation - Bombyx mori (domestic silkworm)
00072 Synthesis and degradation of ketone bodies - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00310 Lysine degradation - Bombyx mori (domestic silkworm)
00380 Tryptophan metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
00650 Butanoate metabolism - Bombyx mori (domestic silkworm)
00900 Terpenoid backbone biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01212 Fatty acid metabolism - Bombyx mori (domestic silkworm)
00072 Synthesis and degradation of ketone bodies - Bombyx mori (domestic silkworm)
00280 Valine, leucine and isoleucine degradation - Bombyx mori (domestic silkworm)
00310 Lysine degradation - Bombyx mori (domestic silkworm)
00380 Tryptophan metabolism - Bombyx mori (domestic silkworm)
00620 Pyruvate metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
00640 Propanoate metabolism - Bombyx mori (domestic silkworm)
00650 Butanoate metabolism - Bombyx mori (domestic silkworm)
00900 Terpenoid backbone biosynthesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01212 Fatty acid metabolism - Bombyx mori (domestic silkworm)
GO
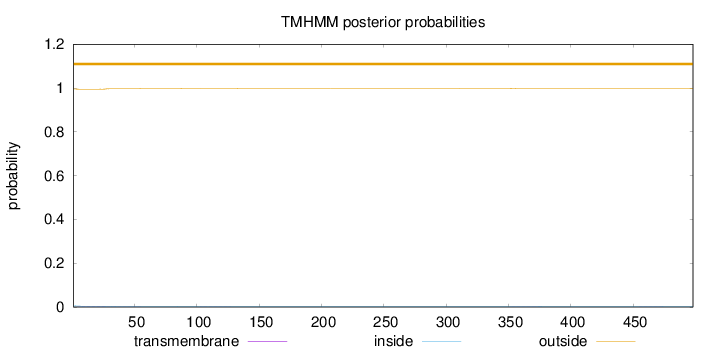
Topology
Length:
498
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0883599999999999
Exp number, first 60 AAs:
0.05946
Total prob of N-in:
0.00492
outside
1 - 498
Population Genetic Test Statistics
Pi
278.925506
Theta
208.942146
Tajima's D
1.168368
CLR
0
CSRT
0.703414829258537
Interpretation
Uncertain
