Gene
KWMTBOMO13666
Pre Gene Modal
BGIBMGA011030
Annotation
PREDICTED:_exportin-4-like_[Bombyx_mori]
Full name
Exportin-4
Location in the cell
PlasmaMembrane Reliability : 1.888
Sequence
CDS
ATGCCGCAATCCGTTTTCTACAATGAGATGTCTCCGATATTGGAATCAGCGTTTGGACGTGGCTCCCGTGGATCATCCTGGGTGGTGTCCCGGCTGGCGGCCCGAGCTGGTACCTGCCTCAGGTACCTGTCCGCTCAGCCGACGGCCGCCTTCCACGCACTCAAACTGCTGACGACATTGGCCCACGCGCACCATAAACAGAATCCTTCGGAGAGCTGCGAGGAGTTCTTGGCGCTGATCGCCTGGGAGGCGGCCGGAAGCAACTTGCCCGGTGAACTGAGGAAAGAATTGCACCGAGCATTCGCAATCGCCGCCACTTACGCTCAGGGAAATGTGAGGACCCGCCTGCTGGACAGCACTGAGGCGCTCCAGAAGAAGTTAATCAACATACTGAACATGGAATCCGACACGGAACCCGTGCGGAGCATGCTCGCCGATACCCTGGACTGCTTCATCGGGGTCACCGAAGGTGTTCTCGAGGTCGGAGCGATGGACGAACAGTTTGTGATGTTAATCGGCGCCTTAGACAAAGTACCGGGTATCTTGTACAGATATCACAACTACCCCGGCGTTGTGTTGCCAGCACTGAATTTGTTATCGAAAAGCGCCAAGAGAATGCTGCACTCGGTTCAACCTCAAAACGTTGATAAGTTCTTAGAAGTATGCAATACTACATTTGAAGTTTATAAACAATGGAATTCTGGAAAGATCTCGAGTATACCACAAGACGCTGAGGATGAAGTGTATGAGGATATATGCGCAATAATGGAACTGATCTGTAGTATAGCGCGGTCAGGAGCCCACACTAACGTTAATGAGACCTGCGCCCGGGGACTGCGACTGTTGCTGCCCTTGATCACTCCTCCGCTGTTGGCACTGCCGTCACTGGCCCAGCACGCATATCGAATGATTAGGGATTTGGATAATTGTGATCAGCTCACAAATCTACCGATAGAGGACTTCAACATGGTGATATCGGCGTTACGCGTCGGGCTGACGGCGGTCTCGTGCGACGTCTCGACGCTGTGCTGCGATACCATCGTGGGCCTGGCGAACAGAGCGCGGACGCTGGGCGACAACAATCCTTACGCCATGTCCTTACTGACGCTGGCCGAACTCTTACTAATGTTGATAATTAAAATGGAAATACCCCCGGACTCGATACCGGCTGCCGGCTCGGCGATATACGCCTTGACGTGCGTGAAGCCGGCTCTCTTAGAGGGCTTAGCCAGACAGCTGATCGAAGCGTACGCGGTCAACGACCCCGCGAACGTGCCCAGATTAGAGGAGGCCTTCAGTCTCCTAACCAACGGTGTACTGTTCGACGGCCTGAGACCTCACAAGATACGCTTCCAAGATAACTTCGATAAGTTCCTAGCCAGCGTTCACGGATTTCTGATTGTCAAATAA
Protein
MPQSVFYNEMSPILESAFGRGSRGSSWVVSRLAARAGTCLRYLSAQPTAAFHALKLLTTLAHAHHKQNPSESCEEFLALIAWEAAGSNLPGELRKELHRAFAIAATYAQGNVRTRLLDSTEALQKKLINILNMESDTEPVRSMLADTLDCFIGVTEGVLEVGAMDEQFVMLIGALDKVPGILYRYHNYPGVVLPALNLLSKSAKRMLHSVQPQNVDKFLEVCNTTFEVYKQWNSGKISSIPQDAEDEVYEDICAIMELICSIARSGAHTNVNETCARGLRLLLPLITPPLLALPSLAQHAYRMIRDLDNCDQLTNLPIEDFNMVISALRVGLTAVSCDVSTLCCDTIVGLANRARTLGDNNPYAMSLLTLAELLLMLIIKMEIPPDSIPAAGSAIYALTCVKPALLEGLARQLIEAYAVNDPANVPRLEEAFSLLTNGVLFDGLRPHKIRFQDNFDKFLASVHGFLIVK
Summary
Description
Mediates the nuclear export of proteins (cargos) with broad substrate specificity. In the nucleus binds cooperatively to its cargo and to the GTPase Ran in its active GTP-bound form. Docking of this trimeric complex to the nuclear pore complex (NPC) is mediated through binding to nucleoporins. Upon transit of a nuclear export complex into the cytoplasm, disassembling of the complex and hydrolysis of Ran-GTP to Ran-GDP (induced by RANBP1 and RANGAP1, respectively) cause release of the cargo from the export receptor. XPO4 then return to the nuclear compartment and mediate another round of transport. The directionality of nuclear export is thought to be conferred by an asymmetric distribution of the GTP- and GDP-bound forms of Ran between the cytoplasm and nucleus (By similarity).
Subunit
Found in a complex with XPO4, Ran and EIF5A. Found in a complex with XPO4, Ran and SMAD3. Interacts with SMAD3. Interacts with Ran and cargo proteins in a GTP-dependent manner (By similarity).
Similarity
Belongs to the exportin family.
Keywords
3D-structure
Complete proteome
Cytoplasm
Nucleus
Phosphoprotein
Protein transport
Reference proteome
Transport
Feature
chain Exportin-4
Uniprot
A0A2A4JXK4
A0A194PN01
A0A2H8TXU3
A0A1B6L6R1
A0A1B6GDH0
U4TTX0
+ More
A0A139W8Z2 A0A2J7PKI5 A0A2S2NWP7 J9JVS3 A0A067QJT8 A0A1B6DTD9 A0A1B6DRC6 A0A2J7PKJ3 A0A1W4XDI0 A0A1Y1LL39 E1ZX91 A0A1Y1LTQ0 A0A1Y1LR33 A0A0V0G7Q3 A0A023EZX8 A0A3L8E119 A0A026WPC1 A0A232EWA4 A0A0P5HI45 K7J792 A0A0P5YUR6 A0A0P5XG18 A0A0P6GGT4 A0A0P5M0Z2 A0A0P5X0Z8 A0A0P6CGU8 E9GHT5 A0A0P5P761 A0A164KE70 A0A0P5EH49 A0A0P6I566 V3ZDV8 A0A0A9W5H9 T1IB11 A0A3P8ZL51 A0A2R5LMJ3 W5UAJ4 A0A2D0R8X3 W5KDP9 A0A146LH98 A0A3B4C0H8 A0A060W0L8 A0A0P5MHY3 E0W318 A0A0R4J254 G3H8I5 Q9ESJ0 A0A1S3NYA4 A0A1S3NYA9 A0A2K6GH78 A0A1A8EQ88 A0A2U9CCM8 H3AB58 A0A061IM28 A0A1A8HGJ4 F6Z8P9 A0A0R4IGB7 A0A1A8K4X0 A0A1A7X4Y7 B0S8I3 A0A1A8AKM7 A0A1A8NK60 A0A1A8LMQ3 A0A1A8R8A1 A0A1A8CQN6 A0A1A8URI1 A0A1A7XZ44 A0A1A8MX12 A0A1A8D7X6 A0A3B3R8S2 A0A1A8PCV0 A0A1A8SDP8 F1RVA6 A0A287AJ95 A0A3B3RA17 A0A0P6DYZ6 A0A3Q0CYP5 A0A2Y9RTG0 A0A1A8JB32 A0A1L8HAY2 Q4R6Q6 G3VUX8 A0A2Y9S296 W5MYC2 A0A1S3NXZ9 W5MYC9
A0A139W8Z2 A0A2J7PKI5 A0A2S2NWP7 J9JVS3 A0A067QJT8 A0A1B6DTD9 A0A1B6DRC6 A0A2J7PKJ3 A0A1W4XDI0 A0A1Y1LL39 E1ZX91 A0A1Y1LTQ0 A0A1Y1LR33 A0A0V0G7Q3 A0A023EZX8 A0A3L8E119 A0A026WPC1 A0A232EWA4 A0A0P5HI45 K7J792 A0A0P5YUR6 A0A0P5XG18 A0A0P6GGT4 A0A0P5M0Z2 A0A0P5X0Z8 A0A0P6CGU8 E9GHT5 A0A0P5P761 A0A164KE70 A0A0P5EH49 A0A0P6I566 V3ZDV8 A0A0A9W5H9 T1IB11 A0A3P8ZL51 A0A2R5LMJ3 W5UAJ4 A0A2D0R8X3 W5KDP9 A0A146LH98 A0A3B4C0H8 A0A060W0L8 A0A0P5MHY3 E0W318 A0A0R4J254 G3H8I5 Q9ESJ0 A0A1S3NYA4 A0A1S3NYA9 A0A2K6GH78 A0A1A8EQ88 A0A2U9CCM8 H3AB58 A0A061IM28 A0A1A8HGJ4 F6Z8P9 A0A0R4IGB7 A0A1A8K4X0 A0A1A7X4Y7 B0S8I3 A0A1A8AKM7 A0A1A8NK60 A0A1A8LMQ3 A0A1A8R8A1 A0A1A8CQN6 A0A1A8URI1 A0A1A7XZ44 A0A1A8MX12 A0A1A8D7X6 A0A3B3R8S2 A0A1A8PCV0 A0A1A8SDP8 F1RVA6 A0A287AJ95 A0A3B3RA17 A0A0P6DYZ6 A0A3Q0CYP5 A0A2Y9RTG0 A0A1A8JB32 A0A1L8HAY2 Q4R6Q6 G3VUX8 A0A2Y9S296 W5MYC2 A0A1S3NXZ9 W5MYC9
Pubmed
26354079
23537049
18362917
19820115
24845553
28004739
+ More
20798317 25474469 30249741 24508170 28648823 20075255 21292972 23254933 25401762 25069045 23127152 25329095 26823975 24755649 20566863 19468303 21183079 21804562 10944119 14621295 9215903 23929341 17495919 23594743 29240929 26319212 28071753 27762356 15944441 21709235
20798317 25474469 30249741 24508170 28648823 20075255 21292972 23254933 25401762 25069045 23127152 25329095 26823975 24755649 20566863 19468303 21183079 21804562 10944119 14621295 9215903 23929341 17495919 23594743 29240929 26319212 28071753 27762356 15944441 21709235
EMBL
NWSH01000423
PCG76496.1
KQ459600
KPI94109.1
GFXV01007320
MBW19125.1
+ More
GEBQ01020627 JAT19350.1 GECZ01009271 JAS60498.1 KB630032 KB631870 ERL83353.1 ERL86848.1 KQ972864 KXZ75751.1 NEVH01024539 PNF16846.1 GGMR01009042 MBY21661.1 ABLF02035477 ABLF02035479 ABLF02035487 KK853274 KDR09117.1 GEDC01008367 JAS28931.1 GEDC01009089 JAS28209.1 PNF16845.1 GEZM01052643 GEZM01052642 JAV74369.1 GL435030 EFN74181.1 GEZM01052644 JAV74367.1 GEZM01052645 JAV74365.1 GECL01001969 JAP04155.1 GBBI01003900 JAC14812.1 QOIP01000001 RLU26404.1 KK107154 EZA56944.1 NNAY01001887 OXU22639.1 GDIQ01229280 JAK22445.1 GDIP01066432 JAM37283.1 GDIP01073643 JAM30072.1 GDIQ01043843 JAN50894.1 GDIQ01173192 JAK78533.1 GDIP01078928 JAM24787.1 GDIP01008821 JAM94894.1 GL732545 EFX80996.1 GDIQ01137010 JAL14716.1 LRGB01003325 KZS03187.1 GDIQ01269764 JAJ81960.1 GDIQ01009415 JAN85322.1 KB202619 ESO89303.1 GBHO01040953 JAG02651.1 ACPB03005775 ACPB03005776 GGLE01006630 MBY10756.1 JT410421 AHH39051.1 GDHC01012769 JAQ05860.1 FR904354 CDQ60596.1 GDIQ01167149 JAK84576.1 DS235881 EEB20024.1 AC154504 AC154675 JH000211 EGV92223.1 AF145021 AK129430 HAEB01002114 SBQ48641.1 CP026256 AWP13536.1 AFYH01134860 AFYH01134861 AFYH01134862 AFYH01134863 AFYH01134864 AFYH01134865 AFYH01134866 AFYH01134867 AFYH01134868 AFYH01134869 KE666079 ERE88314.1 HAEC01015114 SBQ83335.1 CR392366 CU151884 HAEE01006679 SBR26699.1 HADW01011575 SBP12975.1 BX936339 HADY01017104 SBP55589.1 HAEH01002612 SBR69274.1 HAEF01007685 SBR45174.1 HAEH01015252 SBS02261.1 HADZ01018133 SBP82074.1 HAEJ01009764 SBS50221.1 HADX01001114 SBP23346.1 HAEF01020234 SBR61393.1 HAEA01001742 SBQ30222.1 HAEG01007355 SBR79053.1 HAEI01013893 SBS16362.1 AEMK02000075 GDIQ01070583 JAN24154.1 HAED01020424 SBR06980.1 CM004469 OCT93171.1 AB169124 BAE01218.1 AEFK01117146 AEFK01117147 AEFK01117148 AEFK01117149 AHAT01003647 AHAT01003648 AHAT01003649 AHAT01003650 AHAT01003651 AHAT01003652 AHAT01003653
GEBQ01020627 JAT19350.1 GECZ01009271 JAS60498.1 KB630032 KB631870 ERL83353.1 ERL86848.1 KQ972864 KXZ75751.1 NEVH01024539 PNF16846.1 GGMR01009042 MBY21661.1 ABLF02035477 ABLF02035479 ABLF02035487 KK853274 KDR09117.1 GEDC01008367 JAS28931.1 GEDC01009089 JAS28209.1 PNF16845.1 GEZM01052643 GEZM01052642 JAV74369.1 GL435030 EFN74181.1 GEZM01052644 JAV74367.1 GEZM01052645 JAV74365.1 GECL01001969 JAP04155.1 GBBI01003900 JAC14812.1 QOIP01000001 RLU26404.1 KK107154 EZA56944.1 NNAY01001887 OXU22639.1 GDIQ01229280 JAK22445.1 GDIP01066432 JAM37283.1 GDIP01073643 JAM30072.1 GDIQ01043843 JAN50894.1 GDIQ01173192 JAK78533.1 GDIP01078928 JAM24787.1 GDIP01008821 JAM94894.1 GL732545 EFX80996.1 GDIQ01137010 JAL14716.1 LRGB01003325 KZS03187.1 GDIQ01269764 JAJ81960.1 GDIQ01009415 JAN85322.1 KB202619 ESO89303.1 GBHO01040953 JAG02651.1 ACPB03005775 ACPB03005776 GGLE01006630 MBY10756.1 JT410421 AHH39051.1 GDHC01012769 JAQ05860.1 FR904354 CDQ60596.1 GDIQ01167149 JAK84576.1 DS235881 EEB20024.1 AC154504 AC154675 JH000211 EGV92223.1 AF145021 AK129430 HAEB01002114 SBQ48641.1 CP026256 AWP13536.1 AFYH01134860 AFYH01134861 AFYH01134862 AFYH01134863 AFYH01134864 AFYH01134865 AFYH01134866 AFYH01134867 AFYH01134868 AFYH01134869 KE666079 ERE88314.1 HAEC01015114 SBQ83335.1 CR392366 CU151884 HAEE01006679 SBR26699.1 HADW01011575 SBP12975.1 BX936339 HADY01017104 SBP55589.1 HAEH01002612 SBR69274.1 HAEF01007685 SBR45174.1 HAEH01015252 SBS02261.1 HADZ01018133 SBP82074.1 HAEJ01009764 SBS50221.1 HADX01001114 SBP23346.1 HAEF01020234 SBR61393.1 HAEA01001742 SBQ30222.1 HAEG01007355 SBR79053.1 HAEI01013893 SBS16362.1 AEMK02000075 GDIQ01070583 JAN24154.1 HAED01020424 SBR06980.1 CM004469 OCT93171.1 AB169124 BAE01218.1 AEFK01117146 AEFK01117147 AEFK01117148 AEFK01117149 AHAT01003647 AHAT01003648 AHAT01003649 AHAT01003650 AHAT01003651 AHAT01003652 AHAT01003653
Proteomes
UP000218220
UP000053268
UP000030742
UP000007266
UP000235965
UP000007819
+ More
UP000027135 UP000192223 UP000000311 UP000279307 UP000053097 UP000215335 UP000002358 UP000000305 UP000076858 UP000030746 UP000015103 UP000265140 UP000221080 UP000018467 UP000261440 UP000193380 UP000009046 UP000000589 UP000001075 UP000087266 UP000233160 UP000246464 UP000008672 UP000030759 UP000002280 UP000000437 UP000261540 UP000008227 UP000189706 UP000248480 UP000186698 UP000007648 UP000018468
UP000027135 UP000192223 UP000000311 UP000279307 UP000053097 UP000215335 UP000002358 UP000000305 UP000076858 UP000030746 UP000015103 UP000265140 UP000221080 UP000018467 UP000261440 UP000193380 UP000009046 UP000000589 UP000001075 UP000087266 UP000233160 UP000246464 UP000008672 UP000030759 UP000002280 UP000000437 UP000261540 UP000008227 UP000189706 UP000248480 UP000186698 UP000007648 UP000018468
PRIDE
Interpro
SUPFAM
SSF48371
SSF48371
Gene 3D
ProteinModelPortal
A0A2A4JXK4
A0A194PN01
A0A2H8TXU3
A0A1B6L6R1
A0A1B6GDH0
U4TTX0
+ More
A0A139W8Z2 A0A2J7PKI5 A0A2S2NWP7 J9JVS3 A0A067QJT8 A0A1B6DTD9 A0A1B6DRC6 A0A2J7PKJ3 A0A1W4XDI0 A0A1Y1LL39 E1ZX91 A0A1Y1LTQ0 A0A1Y1LR33 A0A0V0G7Q3 A0A023EZX8 A0A3L8E119 A0A026WPC1 A0A232EWA4 A0A0P5HI45 K7J792 A0A0P5YUR6 A0A0P5XG18 A0A0P6GGT4 A0A0P5M0Z2 A0A0P5X0Z8 A0A0P6CGU8 E9GHT5 A0A0P5P761 A0A164KE70 A0A0P5EH49 A0A0P6I566 V3ZDV8 A0A0A9W5H9 T1IB11 A0A3P8ZL51 A0A2R5LMJ3 W5UAJ4 A0A2D0R8X3 W5KDP9 A0A146LH98 A0A3B4C0H8 A0A060W0L8 A0A0P5MHY3 E0W318 A0A0R4J254 G3H8I5 Q9ESJ0 A0A1S3NYA4 A0A1S3NYA9 A0A2K6GH78 A0A1A8EQ88 A0A2U9CCM8 H3AB58 A0A061IM28 A0A1A8HGJ4 F6Z8P9 A0A0R4IGB7 A0A1A8K4X0 A0A1A7X4Y7 B0S8I3 A0A1A8AKM7 A0A1A8NK60 A0A1A8LMQ3 A0A1A8R8A1 A0A1A8CQN6 A0A1A8URI1 A0A1A7XZ44 A0A1A8MX12 A0A1A8D7X6 A0A3B3R8S2 A0A1A8PCV0 A0A1A8SDP8 F1RVA6 A0A287AJ95 A0A3B3RA17 A0A0P6DYZ6 A0A3Q0CYP5 A0A2Y9RTG0 A0A1A8JB32 A0A1L8HAY2 Q4R6Q6 G3VUX8 A0A2Y9S296 W5MYC2 A0A1S3NXZ9 W5MYC9
A0A139W8Z2 A0A2J7PKI5 A0A2S2NWP7 J9JVS3 A0A067QJT8 A0A1B6DTD9 A0A1B6DRC6 A0A2J7PKJ3 A0A1W4XDI0 A0A1Y1LL39 E1ZX91 A0A1Y1LTQ0 A0A1Y1LR33 A0A0V0G7Q3 A0A023EZX8 A0A3L8E119 A0A026WPC1 A0A232EWA4 A0A0P5HI45 K7J792 A0A0P5YUR6 A0A0P5XG18 A0A0P6GGT4 A0A0P5M0Z2 A0A0P5X0Z8 A0A0P6CGU8 E9GHT5 A0A0P5P761 A0A164KE70 A0A0P5EH49 A0A0P6I566 V3ZDV8 A0A0A9W5H9 T1IB11 A0A3P8ZL51 A0A2R5LMJ3 W5UAJ4 A0A2D0R8X3 W5KDP9 A0A146LH98 A0A3B4C0H8 A0A060W0L8 A0A0P5MHY3 E0W318 A0A0R4J254 G3H8I5 Q9ESJ0 A0A1S3NYA4 A0A1S3NYA9 A0A2K6GH78 A0A1A8EQ88 A0A2U9CCM8 H3AB58 A0A061IM28 A0A1A8HGJ4 F6Z8P9 A0A0R4IGB7 A0A1A8K4X0 A0A1A7X4Y7 B0S8I3 A0A1A8AKM7 A0A1A8NK60 A0A1A8LMQ3 A0A1A8R8A1 A0A1A8CQN6 A0A1A8URI1 A0A1A7XZ44 A0A1A8MX12 A0A1A8D7X6 A0A3B3R8S2 A0A1A8PCV0 A0A1A8SDP8 F1RVA6 A0A287AJ95 A0A3B3RA17 A0A0P6DYZ6 A0A3Q0CYP5 A0A2Y9RTG0 A0A1A8JB32 A0A1L8HAY2 Q4R6Q6 G3VUX8 A0A2Y9S296 W5MYC2 A0A1S3NXZ9 W5MYC9
PDB
5DLQ
E-value=6.16063e-17,
Score=215
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Shuttles between the nucleus and the cytoplasm. With evidence from 3 publications.
Nucleus Shuttles between the nucleus and the cytoplasm. With evidence from 3 publications.
Nucleus Shuttles between the nucleus and the cytoplasm. With evidence from 3 publications.
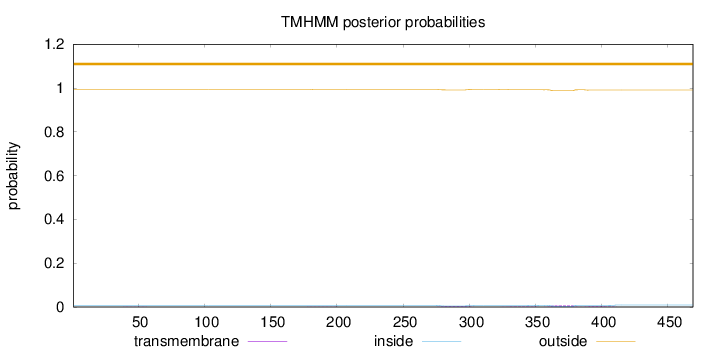
Length:
469
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.351820000000001
Exp number, first 60 AAs:
0.00417
Total prob of N-in:
0.00680
outside
1 - 469
Population Genetic Test Statistics
Pi
310.81928
Theta
197.589434
Tajima's D
2.429942
CLR
0.113494
CSRT
0.935003249837508
Interpretation
Uncertain
