Pre Gene Modal
BGIBMGA011033
Annotation
hypothetical_protein_KGM_21815_[Danaus_plexippus]
Location in the cell
Cytoplasmic Reliability : 1.095 Extracellular Reliability : 1.246
Sequence
CDS
ATGCAGTTATGGCGCCAGGACAGTACAAACACAGGGGTGCTACTCCTACTCTCGTCTATACATTTCCAGTGTCGGAGACTGGACAAGTCTGCTCATTTTTCCACATTGGCGATCAAACAGAATCCTTTGTTGGCTGAAGCCTACAGCAACTTGGGCAATGTTTACAAAGAACGCGGGCAGCTTCAGGAGGCGCTGGAGAACTATCGTCACGCTGTCCGTTTGAAGCCGGACTTCATCGATGGATACATCAACTTGGCTGCGGCTCTCGTTGCCGCCGGCGATATGGAGCAAGCTGTGCAGGCTTACGTTACCGCACTGCAGTATAACCCTGATTTGTACTGCGTCAGAAGCGATTTGGGCAATTTGCTCAAAGCCCTCGGAAGACTAGACGAGGCAAAGGCTTGTTACTTGAAGGCAATCGAAACTAGACCCGATTTCGCGGTCGCCTGGAGCAACTTGGGTTGCGTTTTCAATGCGCAGAGTGAAATATGGTTGGCAATTCATCATTTCGAAAAAGCTGTCGCATTGGATCCCAATTTCTTAGACGCGTACATTAATTTAGGAAATGTACTAAAAGAAGCTAGAATTTTTGACAGGGCCGTCGCCGCATATTTGAGAGCCTTGAATTTATCTCCCAATAATGCCGTCGTGCACGGCAATTTGGCCTGCGTGTACTACGAACAAGGACTGATTGACTTGGCCATCGATACTTATCGACGTGCCATCGAATTACAACAGAACTTCCCGGACGCGTATTGCAATTTAGCTAATGCACTCAAAGAAAAGGGACAAGTGACTGATGCTGAAGAGTGTTATAATACCGCGTTACGTCTCTGTCCGTCTCATGCAGACTCCCTCAACAATCTGGCTAATATAAAACGGGAACAAGGCTACATCGAGGAGGCTACACGTTTATATTTAAAAGCATTAGAAGTGTTCCCGGAGTTTGCGGCGGCGCATAGTAATCTGGCTTCAGTGTTGCAACAACAAGGGAAACTAAACGAAGCCCTTATGCATTACAAGGAAGCTATACGTATTCAGCCGACCTTTGCCGATGCGTATAGCAACATGGGCAATACGCTGAAGGAAATGCAAGATGTAGCCGGTGCTCTGCAATGCTACACAAGGGCGATTCAAATTAATCCCGCATTTGCGGATGCTCATAGCAATTTGGCCAGCATCCACAAAGATTCTGGTAACATTCCTGAAGCTATACAGTCGTACAGAACAGCGCTTAAGCTTAAACCAGACTTCCCAGATGCGTATTGCAATTTAGCCCACTGTTTACAAATAGTGTGTGACTGGACGGATTATGAGGCTCGAATGAAAAAATTGGTCAGCATTGTCGCTGAGCAGCTGGAGAAAAACAGACTGCCATCAGTACATCCGCACCACTCCATGTTGTATCCGCTGACACATGACTTTAGGAAGGCCATCGCCGCCCGTCACGCGAACCTTTGCTTGGAGAAAGTGCAAGTGTTGCACAAAAACCCTTACAAGTTCCCTAGAGATTTACAAGGCCGCCTTCGTATCGGGTACGTAAGCAGCGATTTCGGTAACCATCCGACATCGCATTTGATGCAATCGGTACCTGGTTTGCATGACCGAACAAAAGTGGAAATATTCTGCTATGCCCTCAGTCCGGACGACGGAACAACGTTCCGATCGAAAATCGCCAGGGAAGCGGAGCATTTTATCGATCTATCTCAGATGCCTTGTAACGGCAAAGCAGCCGATAAAATATACGGAGACGGTATTCATATCTTGGTCAATATGAATGGATACACTAAAGGAGCGCGAAATGAAATTTTCGCTTTGAGACCAGCCCCGGTCCAGGTAATGTGGCTCGGGTACCCGGGAACTAGCGGGGCTAGTTACATGGACTATTTAGTTACTGATGCAGTGACGTCACCCGTGGAACTTGCCAGCCAATATAGCGAAAAACTAGCGTACATGCCACACACGTATTTTGTTGGCGATCACAAGCAAATGTTCCCACATTTACAAGAACGTCTGATTCTGAGCGACAAAGTTAAATCCCACAACAATTTGGGAAGTGTCGCGGACAATGTTGCTGTGATTAACGCGACCGATCTGTCGCCCCTTGTGGAAAATACTGACATTAAAGAAATAAAAGAGATCGTGCGAGCAGCCAGACCCGTTGAAATATCTTTGAAAGTAGCCGAATTGCCAACGACGACACCCATTGAAACTATGATTGCATCAGGACAAGTACAGACATCCGTAAATGGCGTTATTCTTCAAAATGGCTTAGCTACTACGCAGACGAACAACAAGGCTGCGACGGGCGAGGAAGTACCACAATCTATAGTGATTACAACTCGACAACAATACGGCTTACCGGATGACGCCGTAGTGTACTGCAACTTCAACCAATTGTATAAGATTGACCCATTAACTCTACACATGTGGGTGTACATACTGAAACATGTACCTAACAGTGTATTATGGTTATTAAGATTCCCGGCCGTAGGAGAACCCAATTTACAATCCACTGCCAACCAACTTGGTCTTCCGCCGGGACGGATAATATTCTCAAATGTTGCTGCCAAAGAAGAGCACGTACGACGCGGACAGTTAGCCGATGTGTGTCTCGACACTCCACTTTGTAATGGACACACAACTAGTATGGACATCCTATGGACTGGCACACCGGTTGTGACCCTTCCTGGGGAGACTTTGGCGTCTCGAGTGGCAGCTTCTCAATTAAATACTTTAGGATGTCCCGAGCTCATTGCCAGAACGAGACAAGAATACCAAGACATAGCTGTAAGACTGGGAACAGATAGAGAATACCTAAAAGCGATCAGAGCAAAGGTGTGGACAGCTCGCACTGACAGTCCTCTTTTCGACTGCAAGGCGTACGCCACCGGACTTGAGATGCTGTACAACAGGATGTGGTCGAGGCACGCGCGAGGCGATCGCCCTGATCATATACAGGCCCTCGAGAAATAA
Protein
MQLWRQDSTNTGVLLLLSSIHFQCRRLDKSAHFSTLAIKQNPLLAEAYSNLGNVYKERGQLQEALENYRHAVRLKPDFIDGYINLAAALVAAGDMEQAVQAYVTALQYNPDLYCVRSDLGNLLKALGRLDEAKACYLKAIETRPDFAVAWSNLGCVFNAQSEIWLAIHHFEKAVALDPNFLDAYINLGNVLKEARIFDRAVAAYLRALNLSPNNAVVHGNLACVYYEQGLIDLAIDTYRRAIELQQNFPDAYCNLANALKEKGQVTDAEECYNTALRLCPSHADSLNNLANIKREQGYIEEATRLYLKALEVFPEFAAAHSNLASVLQQQGKLNEALMHYKEAIRIQPTFADAYSNMGNTLKEMQDVAGALQCYTRAIQINPAFADAHSNLASIHKDSGNIPEAIQSYRTALKLKPDFPDAYCNLAHCLQIVCDWTDYEARMKKLVSIVAEQLEKNRLPSVHPHHSMLYPLTHDFRKAIAARHANLCLEKVQVLHKNPYKFPRDLQGRLRIGYVSSDFGNHPTSHLMQSVPGLHDRTKVEIFCYALSPDDGTTFRSKIAREAEHFIDLSQMPCNGKAADKIYGDGIHILVNMNGYTKGARNEIFALRPAPVQVMWLGYPGTSGASYMDYLVTDAVTSPVELASQYSEKLAYMPHTYFVGDHKQMFPHLQERLILSDKVKSHNNLGSVADNVAVINATDLSPLVENTDIKEIKEIVRAARPVEISLKVAELPTTTPIETMIASGQVQTSVNGVILQNGLATTQTNNKAATGEEVPQSIVITTRQQYGLPDDAVVYCNFNQLYKIDPLTLHMWVYILKHVPNSVLWLLRFPAVGEPNLQSTANQLGLPPGRIIFSNVAAKEEHVRRGQLADVCLDTPLCNGHTTSMDILWTGTPVVTLPGETLASRVAASQLNTLGCPELIARTRQEYQDIAVRLGTDREYLKAIRAKVWTARTDSPLFDCKAYATGLEMLYNRMWSRHARGDRPDHIQALEK
Summary
Catalytic Activity
S-ubiquitinyl-[E2 ubiquitin-conjugating enzyme]-L-cysteine + [acceptor protein]-L-lysine = [E2 ubiquitin-conjugating enzyme]-L-cysteine + N(6)-ubiquitinyl-[acceptor protein]-L-lysine.
Uniprot
H9JND0
A0A212ETF9
A0A2H1VIU1
A0A194PPU3
A0A194R9U0
A0A2A4JXZ0
+ More
A0A1L8E217 D6WHK3 A0A1L8E1M7 A0A2A3EG57 A0A1Y1N3F7 V9IAZ6 V9I8Q6 A0A1Y1N648 A0A0T6B1V8 A0A195EWP2 A0A151HYK7 A0A0M9A6T3 A0A158NSR3 F4W877 A0A3L8D9Q0 A0A026WWW3 A0A310SJY3 A0A182J0E2 A0A182LLH0 E2AQU9 A0A182YPW9 Q7Q5R0 A0A2M4A4P0 A0A2M4CTZ1 A0A2M4A5I3 A0A1B6JPC7 A0A182QWV3 A0A0P4VNF7 A0A023F5R9 A0A069DZQ2 A0A2M4BB22 A0A2M4BBG1 A0A2M4BBC0 A0A2M4CV31 A0A2M3YZF6 A0A0V0G3N5 A0A1Q3FUS6 A0A0A9Z4Y3 A0A0A9X5K8 U5ETN0 A0A224XDN0 E0W1P3 U4UA05 B4MQS7 A0A0M5J365 N6TQB0 A0A3B0K6S0 A0A336MKK1 B4KLX3 A0A1I8NQD3 A0A1I8NQD1 A0A1A9UJ03 A0A1B0FFJ6 A0A1I8NBP2 A0A1I8NQC9 A0A1B0A991 A0A1I8NBP7 A0A1B0BI91 A0A1A9WP55 A0A1I8NBN1 W8AXT6 T1P8Z7 A0A0J9R6G6 Q7KJA9 A0A0R1E7G4 A0A0Q5WP95 B4LP13 B4J676 A0A034WVC4 A0A0A1WLG3 A0A0K8WCN3 A0A0L7L8G3 X1WJ62 A0A3Q0IVN3 A0A087UUV5 A0A1W7RB52 E9H0Z0 A0A0P5TS64 A0A0P5N6U8 A0A0P6B764 A0A0P6D1Z6 A0A3R7SIH9 Q9Y148 A0A1Z5L5B0 A0A0P4VSK1 A0A293M115 A0A2R5LP27 A0A023FNI2 A0A131Z5W0 A0A023FVE7 T1JCD7 A0A224YX38 V5I096 A0A0L0CHP1
A0A1L8E217 D6WHK3 A0A1L8E1M7 A0A2A3EG57 A0A1Y1N3F7 V9IAZ6 V9I8Q6 A0A1Y1N648 A0A0T6B1V8 A0A195EWP2 A0A151HYK7 A0A0M9A6T3 A0A158NSR3 F4W877 A0A3L8D9Q0 A0A026WWW3 A0A310SJY3 A0A182J0E2 A0A182LLH0 E2AQU9 A0A182YPW9 Q7Q5R0 A0A2M4A4P0 A0A2M4CTZ1 A0A2M4A5I3 A0A1B6JPC7 A0A182QWV3 A0A0P4VNF7 A0A023F5R9 A0A069DZQ2 A0A2M4BB22 A0A2M4BBG1 A0A2M4BBC0 A0A2M4CV31 A0A2M3YZF6 A0A0V0G3N5 A0A1Q3FUS6 A0A0A9Z4Y3 A0A0A9X5K8 U5ETN0 A0A224XDN0 E0W1P3 U4UA05 B4MQS7 A0A0M5J365 N6TQB0 A0A3B0K6S0 A0A336MKK1 B4KLX3 A0A1I8NQD3 A0A1I8NQD1 A0A1A9UJ03 A0A1B0FFJ6 A0A1I8NBP2 A0A1I8NQC9 A0A1B0A991 A0A1I8NBP7 A0A1B0BI91 A0A1A9WP55 A0A1I8NBN1 W8AXT6 T1P8Z7 A0A0J9R6G6 Q7KJA9 A0A0R1E7G4 A0A0Q5WP95 B4LP13 B4J676 A0A034WVC4 A0A0A1WLG3 A0A0K8WCN3 A0A0L7L8G3 X1WJ62 A0A3Q0IVN3 A0A087UUV5 A0A1W7RB52 E9H0Z0 A0A0P5TS64 A0A0P5N6U8 A0A0P6B764 A0A0P6D1Z6 A0A3R7SIH9 Q9Y148 A0A1Z5L5B0 A0A0P4VSK1 A0A293M115 A0A2R5LP27 A0A023FNI2 A0A131Z5W0 A0A023FVE7 T1JCD7 A0A224YX38 V5I096 A0A0L0CHP1
Pubmed
19121390
22118469
26354079
18362917
19820115
28004739
+ More
21347285 21719571 30249741 24508170 20966253 20798317 25244985 12364791 14747013 17210077 27129103 25474469 26334808 25401762 26823975 20566863 23537049 17994087 25315136 24495485 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26674417 17550304 18057021 25348373 25830018 26227816 21292972 28528879 26830274 28797301 25765539 26108605
21347285 21719571 30249741 24508170 20966253 20798317 25244985 12364791 14747013 17210077 27129103 25474469 26334808 25401762 26823975 20566863 23537049 17994087 25315136 24495485 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26674417 17550304 18057021 25348373 25830018 26227816 21292972 28528879 26830274 28797301 25765539 26108605
EMBL
BABH01019450
AGBW02012603
OWR44731.1
ODYU01002802
SOQ40748.1
KQ459597
+ More
KPI94754.1 KQ460473 KPJ14407.1 NWSH01000423 PCG76494.1 GFDF01001462 JAV12622.1 KQ971331 EFA01003.1 GFDF01001473 JAV12611.1 KZ288255 PBC30698.1 GEZM01013689 GEZM01013687 JAV92369.1 JR036953 AEY57499.1 JR036952 AEY57498.1 GEZM01013688 JAV92370.1 LJIG01016195 KRT81330.1 KQ981948 KYN32648.1 KQ976723 KYM76681.1 KQ435724 KOX78410.1 ADTU01025121 GL887898 EGI69572.1 QOIP01000011 RLU16638.1 KK107078 EZA60211.1 KQ760514 OAD60022.1 GL441834 EFN64211.1 AAAB01008960 EAA10760.4 GGFK01002408 MBW35729.1 GGFL01004463 MBW68641.1 GGFK01002651 MBW35972.1 GECU01006701 JAT01006.1 AXCN02000346 GDKW01000423 JAI56172.1 GBBI01002194 JAC16518.1 GBGD01000245 JAC88644.1 GGFJ01001095 MBW50236.1 GGFJ01001238 MBW50379.1 GGFJ01001195 MBW50336.1 GGFL01005015 MBW69193.1 GGFM01000894 MBW21645.1 GECL01003455 JAP02669.1 GFDL01003711 JAV31334.1 GBHO01003282 GDHC01006945 JAG40322.1 JAQ11684.1 GBHO01027602 JAG16002.1 GANO01001840 JAB58031.1 GFTR01008528 JAW07898.1 DS235873 EEB19625.1 KB630245 KB630279 KB631948 ERL83496.1 ERL83514.1 ERL87411.1 CH963849 EDW74466.1 CP012524 ALC42656.1 APGK01058526 KB741291 ENN70496.1 OUUW01000032 SPP89814.1 UFQT01001415 SSX30450.1 CH933808 EDW09783.1 CCAG010016602 JXJN01014862 GAMC01012750 JAB93805.1 KA645251 AFP59880.1 CM002911 KMY91610.1 KMY91611.1 AF217788 AE013599 BT032865 AAF32311.1 AAF57338.1 AAG22338.1 AAG22339.1 ACD81879.1 CH891608 KRK05133.1 CH954177 KQS71055.1 CH940648 EDW61182.1 KRF79836.1 KRF79837.1 CH916367 EDW01934.1 GAKP01001254 JAC57698.1 GBXI01014423 JAC99868.1 GDHF01023546 GDHF01003699 JAI28768.1 JAI48615.1 JTDY01002347 KOB71619.1 ABLF02036676 KK121753 KFM81144.1 GFAH01000023 JAV48366.1 GL732582 EFX74619.1 GDIQ01253079 GDIQ01221586 GDIQ01142217 GDIP01123524 GDIQ01010608 LRGB01003134 JAJ98645.1 JAL80190.1 KZS04078.1 GDIQ01146514 JAL05212.1 GDIP01033088 JAM70627.1 GDIQ01084085 JAN10652.1 QCYY01003673 ROT62470.1 AF145622 AAD38597.1 GFJQ02004367 JAW02603.1 GDRN01100857 JAI58476.1 GFWV01009277 MAA34006.1 GGLE01007113 MBY11239.1 GBBK01002057 JAC22425.1 GEDV01003056 JAP85501.1 GBBL01002597 JAC24723.1 JH432064 GFPF01011031 MAA22177.1 GANP01003940 JAB80528.1 JRES01000466 KNC31004.1
KPI94754.1 KQ460473 KPJ14407.1 NWSH01000423 PCG76494.1 GFDF01001462 JAV12622.1 KQ971331 EFA01003.1 GFDF01001473 JAV12611.1 KZ288255 PBC30698.1 GEZM01013689 GEZM01013687 JAV92369.1 JR036953 AEY57499.1 JR036952 AEY57498.1 GEZM01013688 JAV92370.1 LJIG01016195 KRT81330.1 KQ981948 KYN32648.1 KQ976723 KYM76681.1 KQ435724 KOX78410.1 ADTU01025121 GL887898 EGI69572.1 QOIP01000011 RLU16638.1 KK107078 EZA60211.1 KQ760514 OAD60022.1 GL441834 EFN64211.1 AAAB01008960 EAA10760.4 GGFK01002408 MBW35729.1 GGFL01004463 MBW68641.1 GGFK01002651 MBW35972.1 GECU01006701 JAT01006.1 AXCN02000346 GDKW01000423 JAI56172.1 GBBI01002194 JAC16518.1 GBGD01000245 JAC88644.1 GGFJ01001095 MBW50236.1 GGFJ01001238 MBW50379.1 GGFJ01001195 MBW50336.1 GGFL01005015 MBW69193.1 GGFM01000894 MBW21645.1 GECL01003455 JAP02669.1 GFDL01003711 JAV31334.1 GBHO01003282 GDHC01006945 JAG40322.1 JAQ11684.1 GBHO01027602 JAG16002.1 GANO01001840 JAB58031.1 GFTR01008528 JAW07898.1 DS235873 EEB19625.1 KB630245 KB630279 KB631948 ERL83496.1 ERL83514.1 ERL87411.1 CH963849 EDW74466.1 CP012524 ALC42656.1 APGK01058526 KB741291 ENN70496.1 OUUW01000032 SPP89814.1 UFQT01001415 SSX30450.1 CH933808 EDW09783.1 CCAG010016602 JXJN01014862 GAMC01012750 JAB93805.1 KA645251 AFP59880.1 CM002911 KMY91610.1 KMY91611.1 AF217788 AE013599 BT032865 AAF32311.1 AAF57338.1 AAG22338.1 AAG22339.1 ACD81879.1 CH891608 KRK05133.1 CH954177 KQS71055.1 CH940648 EDW61182.1 KRF79836.1 KRF79837.1 CH916367 EDW01934.1 GAKP01001254 JAC57698.1 GBXI01014423 JAC99868.1 GDHF01023546 GDHF01003699 JAI28768.1 JAI48615.1 JTDY01002347 KOB71619.1 ABLF02036676 KK121753 KFM81144.1 GFAH01000023 JAV48366.1 GL732582 EFX74619.1 GDIQ01253079 GDIQ01221586 GDIQ01142217 GDIP01123524 GDIQ01010608 LRGB01003134 JAJ98645.1 JAL80190.1 KZS04078.1 GDIQ01146514 JAL05212.1 GDIP01033088 JAM70627.1 GDIQ01084085 JAN10652.1 QCYY01003673 ROT62470.1 AF145622 AAD38597.1 GFJQ02004367 JAW02603.1 GDRN01100857 JAI58476.1 GFWV01009277 MAA34006.1 GGLE01007113 MBY11239.1 GBBK01002057 JAC22425.1 GEDV01003056 JAP85501.1 GBBL01002597 JAC24723.1 JH432064 GFPF01011031 MAA22177.1 GANP01003940 JAB80528.1 JRES01000466 KNC31004.1
Proteomes
UP000005204
UP000007151
UP000053268
UP000053240
UP000218220
UP000007266
+ More
UP000242457 UP000078541 UP000078540 UP000053105 UP000005205 UP000007755 UP000279307 UP000053097 UP000075880 UP000075882 UP000000311 UP000076408 UP000007062 UP000075886 UP000009046 UP000030742 UP000007798 UP000092553 UP000019118 UP000268350 UP000009192 UP000095300 UP000078200 UP000092444 UP000095301 UP000092445 UP000092460 UP000091820 UP000000803 UP000002282 UP000008711 UP000008792 UP000001070 UP000037510 UP000007819 UP000079169 UP000054359 UP000000305 UP000076858 UP000283509 UP000037069
UP000242457 UP000078541 UP000078540 UP000053105 UP000005205 UP000007755 UP000279307 UP000053097 UP000075880 UP000075882 UP000000311 UP000076408 UP000007062 UP000075886 UP000009046 UP000030742 UP000007798 UP000092553 UP000019118 UP000268350 UP000009192 UP000095300 UP000078200 UP000092444 UP000095301 UP000092445 UP000092460 UP000091820 UP000000803 UP000002282 UP000008711 UP000008792 UP000001070 UP000037510 UP000007819 UP000079169 UP000054359 UP000000305 UP000076858 UP000283509 UP000037069
Pfam
Interpro
IPR013026
TPR-contain_dom
+ More
IPR037919 OGT
IPR011990 TPR-like_helical_dom_sf
IPR019734 TPR_repeat
IPR029489 OGT/SEC/SPY_C
IPR001440 TPR_1
IPR003107 HAT
IPR000569 HECT_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR035983 Hect_E3_ubiquitin_ligase
IPR001680 WD40_repeat
IPR035892 C2_domain_sf
IPR017986 WD40_repeat_dom
IPR020472 G-protein_beta_WD-40_rep
IPR036020 WW_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR001202 WW_dom
IPR000008 C2_dom
IPR000077 Ribosomal_L39
IPR020083 Ribosomal_L39_CS
IPR023626 Ribosomal_L39e_dom_sf
IPR013105 TPR_2
IPR037919 OGT
IPR011990 TPR-like_helical_dom_sf
IPR019734 TPR_repeat
IPR029489 OGT/SEC/SPY_C
IPR001440 TPR_1
IPR003107 HAT
IPR000569 HECT_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR035983 Hect_E3_ubiquitin_ligase
IPR001680 WD40_repeat
IPR035892 C2_domain_sf
IPR017986 WD40_repeat_dom
IPR020472 G-protein_beta_WD-40_rep
IPR036020 WW_dom_sf
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR001202 WW_dom
IPR000008 C2_dom
IPR000077 Ribosomal_L39
IPR020083 Ribosomal_L39_CS
IPR023626 Ribosomal_L39e_dom_sf
IPR013105 TPR_2
SUPFAM
Gene 3D
ProteinModelPortal
H9JND0
A0A212ETF9
A0A2H1VIU1
A0A194PPU3
A0A194R9U0
A0A2A4JXZ0
+ More
A0A1L8E217 D6WHK3 A0A1L8E1M7 A0A2A3EG57 A0A1Y1N3F7 V9IAZ6 V9I8Q6 A0A1Y1N648 A0A0T6B1V8 A0A195EWP2 A0A151HYK7 A0A0M9A6T3 A0A158NSR3 F4W877 A0A3L8D9Q0 A0A026WWW3 A0A310SJY3 A0A182J0E2 A0A182LLH0 E2AQU9 A0A182YPW9 Q7Q5R0 A0A2M4A4P0 A0A2M4CTZ1 A0A2M4A5I3 A0A1B6JPC7 A0A182QWV3 A0A0P4VNF7 A0A023F5R9 A0A069DZQ2 A0A2M4BB22 A0A2M4BBG1 A0A2M4BBC0 A0A2M4CV31 A0A2M3YZF6 A0A0V0G3N5 A0A1Q3FUS6 A0A0A9Z4Y3 A0A0A9X5K8 U5ETN0 A0A224XDN0 E0W1P3 U4UA05 B4MQS7 A0A0M5J365 N6TQB0 A0A3B0K6S0 A0A336MKK1 B4KLX3 A0A1I8NQD3 A0A1I8NQD1 A0A1A9UJ03 A0A1B0FFJ6 A0A1I8NBP2 A0A1I8NQC9 A0A1B0A991 A0A1I8NBP7 A0A1B0BI91 A0A1A9WP55 A0A1I8NBN1 W8AXT6 T1P8Z7 A0A0J9R6G6 Q7KJA9 A0A0R1E7G4 A0A0Q5WP95 B4LP13 B4J676 A0A034WVC4 A0A0A1WLG3 A0A0K8WCN3 A0A0L7L8G3 X1WJ62 A0A3Q0IVN3 A0A087UUV5 A0A1W7RB52 E9H0Z0 A0A0P5TS64 A0A0P5N6U8 A0A0P6B764 A0A0P6D1Z6 A0A3R7SIH9 Q9Y148 A0A1Z5L5B0 A0A0P4VSK1 A0A293M115 A0A2R5LP27 A0A023FNI2 A0A131Z5W0 A0A023FVE7 T1JCD7 A0A224YX38 V5I096 A0A0L0CHP1
A0A1L8E217 D6WHK3 A0A1L8E1M7 A0A2A3EG57 A0A1Y1N3F7 V9IAZ6 V9I8Q6 A0A1Y1N648 A0A0T6B1V8 A0A195EWP2 A0A151HYK7 A0A0M9A6T3 A0A158NSR3 F4W877 A0A3L8D9Q0 A0A026WWW3 A0A310SJY3 A0A182J0E2 A0A182LLH0 E2AQU9 A0A182YPW9 Q7Q5R0 A0A2M4A4P0 A0A2M4CTZ1 A0A2M4A5I3 A0A1B6JPC7 A0A182QWV3 A0A0P4VNF7 A0A023F5R9 A0A069DZQ2 A0A2M4BB22 A0A2M4BBG1 A0A2M4BBC0 A0A2M4CV31 A0A2M3YZF6 A0A0V0G3N5 A0A1Q3FUS6 A0A0A9Z4Y3 A0A0A9X5K8 U5ETN0 A0A224XDN0 E0W1P3 U4UA05 B4MQS7 A0A0M5J365 N6TQB0 A0A3B0K6S0 A0A336MKK1 B4KLX3 A0A1I8NQD3 A0A1I8NQD1 A0A1A9UJ03 A0A1B0FFJ6 A0A1I8NBP2 A0A1I8NQC9 A0A1B0A991 A0A1I8NBP7 A0A1B0BI91 A0A1A9WP55 A0A1I8NBN1 W8AXT6 T1P8Z7 A0A0J9R6G6 Q7KJA9 A0A0R1E7G4 A0A0Q5WP95 B4LP13 B4J676 A0A034WVC4 A0A0A1WLG3 A0A0K8WCN3 A0A0L7L8G3 X1WJ62 A0A3Q0IVN3 A0A087UUV5 A0A1W7RB52 E9H0Z0 A0A0P5TS64 A0A0P5N6U8 A0A0P6B764 A0A0P6D1Z6 A0A3R7SIH9 Q9Y148 A0A1Z5L5B0 A0A0P4VSK1 A0A293M115 A0A2R5LP27 A0A023FNI2 A0A131Z5W0 A0A023FVE7 T1JCD7 A0A224YX38 V5I096 A0A0L0CHP1
PDB
5A01
E-value=0,
Score=3282
Ontologies
GO
PANTHER
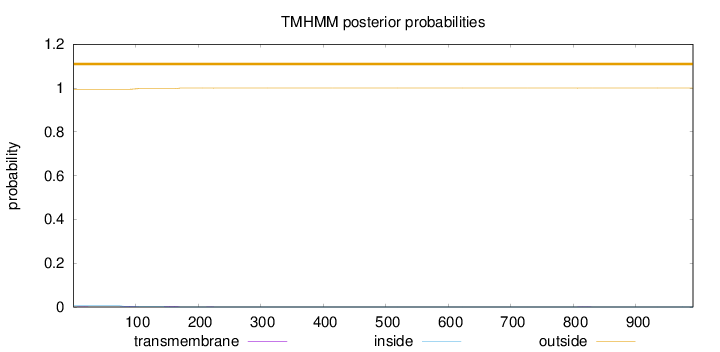
Topology
Length:
991
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.12309
Exp number, first 60 AAs:
0.00542
Total prob of N-in:
0.00555
outside
1 - 991
Population Genetic Test Statistics
Pi
300.014716
Theta
205.997332
Tajima's D
1.478554
CLR
0.372515
CSRT
0.782310884455777
Interpretation
Uncertain
