Gene
KWMTBOMO13654
Pre Gene Modal
BGIBMGA011077
Annotation
PREDICTED:_circadian_clock-controlled_protein-like_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 2.369
Sequence
CDS
ATGCTTACTACACTGTTACTATTATCATGTGCATTTTTCAGTGCTTCGGGTGAAAAAGAAAAATGCAAAGTTACAGACAAGCAATGTATTCTTTTGCTCAGCAATTCTGTCTTTGAAAAAGTATTCGAAGCTGATAATAATCAAAACGTCGATCCCATGTACATAGAGTTCATGGAAGGAAATTTTCTTGACCTGAAGATCAAGTTCAGAAATATTTCCATGACTGGCTATAATACCTGTAAAGTCTTCGATTTATATTGGGATATAGAACAATTTTTATTCAACTTCGAATTGTACTGTTCGAGAATTTCGATTAATGGTCAATATGAGATATCTGGACCGGCTGTGACAACTGAAGAAAAAGGAAAATATACTATTAATACAAAAAGCTACAAGTTAATGACGAACACGACATTTGAGCTTGTACAAACTGTTGGCAATAAAACTGTTTTTCATGTTAAGAACTTCAACGCCAAGGTGTCGCCGTTGGAAAAAGTGGCTTTACATTTCACGAATCTTTATAACGGGCAGGAAATACCAAGTGCCAAATTTTTAACACAAGAGCACTGGAGAAATATTATCTACACTCTTCAAGATAACTTCGCTACAATCTGCTTCAGGAATTTATTTCTAGCTTTAAATAAACTATTTACAACATTGCCATTAGAAGAATATTTGGAATTGTAG
Protein
MLTTLLLLSCAFFSASGEKEKCKVTDKQCILLLSNSVFEKVFEADNNQNVDPMYIEFMEGNFLDLKIKFRNISMTGYNTCKVFDLYWDIEQFLFNFELYCSRISINGQYEISGPAVTTEEKGKYTINTKSYKLMTNTTFELVQTVGNKTVFHVKNFNAKVSPLEKVALHFTNLYNGQEIPSAKFLTQEHWRNIIYTLQDNFATICFRNLFLALNKLFTTLPLEEYLEL
Summary
Uniprot
D2SNL3
A0A3B0EWA9
A0A3G1KLC0
A0A3B0ET75
A0A2H1WKM4
A0A194PR79
+ More
A0A212F1C0 A0A194REN7 A0A1E1WUL1 A0A2H1V0V6 A0A0K8TVA7 H9IVP4 Q402D8 D2SNW8 A0A3G1KKW8 A0A2H1V896 A0A2W1BMT1 Q86GJ4 A0A2A4JM86 A0A2A4JKV9 A0A3B0EWC7 A0A2W1BVK0 A0A2H1V876 A0A161IWD8 A0A0P0EUL4 A0A3S2P7X1 A0A2H1WKZ8 D2SNM7 A0A0N0PDH3 A0A0L7LG13 A0A1A9YIN2 A0A1B0AWP1 A0A2H1V2G1 I4DMS7 D2SNV5 A0A3G1KL03 A0A1E1W258 A0A2W1BMF2 D2SNY3 I4DKQ2 A0A212F1D1 A0A2H1WF65 A0A2A4JHG1 A0A2J7QTH9 A0A194QA85 A0A168T0I5 A0A3G1NI93 A0A2J7QTH3 A0A0K8TU33 A0A3B0EYT6 A0A3G1KKZ9 A0A0K8TVA3 A0A3S2M186 A0A194RFL6 S4NQ33 A0A2A4JGK7 A0A2H1W5H9 A0A2J7QTJ5 A0A2A4JN36 A0A2R7WZQ2 A0A3B0JTT6 A0A3G1KKW6 A0A3B0EU73 A0A2J7QTH5 A0A194RAT9 H9JNH2 A0A1B0A461 A0A2K8GKQ0 A0A2H1VDE1 A0A161JCG7 Q402E0 C6KYM6 A0A0A9YNG7 A0A0K8TUQ8 A0A2A4JLY9 A0A2H1X2K5 A0A3B0JS36 A0A0P0EUK7 A0A2A4J606 A0A0K8TU38 A0A212EXB5 A0A2H1VI77 A0A2H1VVY8 A0A212FJZ7 A0A212F9C7 A0A336MJ36 A0A067R2U4 A0A2H1VI74 U4URM8 R4V1J3 N6U7G6 A0A1W4WTJ1 A0A1I8NVH9 A0A1I8MK16 A0A1A9VQQ0
A0A212F1C0 A0A194REN7 A0A1E1WUL1 A0A2H1V0V6 A0A0K8TVA7 H9IVP4 Q402D8 D2SNW8 A0A3G1KKW8 A0A2H1V896 A0A2W1BMT1 Q86GJ4 A0A2A4JM86 A0A2A4JKV9 A0A3B0EWC7 A0A2W1BVK0 A0A2H1V876 A0A161IWD8 A0A0P0EUL4 A0A3S2P7X1 A0A2H1WKZ8 D2SNM7 A0A0N0PDH3 A0A0L7LG13 A0A1A9YIN2 A0A1B0AWP1 A0A2H1V2G1 I4DMS7 D2SNV5 A0A3G1KL03 A0A1E1W258 A0A2W1BMF2 D2SNY3 I4DKQ2 A0A212F1D1 A0A2H1WF65 A0A2A4JHG1 A0A2J7QTH9 A0A194QA85 A0A168T0I5 A0A3G1NI93 A0A2J7QTH3 A0A0K8TU33 A0A3B0EYT6 A0A3G1KKZ9 A0A0K8TVA3 A0A3S2M186 A0A194RFL6 S4NQ33 A0A2A4JGK7 A0A2H1W5H9 A0A2J7QTJ5 A0A2A4JN36 A0A2R7WZQ2 A0A3B0JTT6 A0A3G1KKW6 A0A3B0EU73 A0A2J7QTH5 A0A194RAT9 H9JNH2 A0A1B0A461 A0A2K8GKQ0 A0A2H1VDE1 A0A161JCG7 Q402E0 C6KYM6 A0A0A9YNG7 A0A0K8TUQ8 A0A2A4JLY9 A0A2H1X2K5 A0A3B0JS36 A0A0P0EUK7 A0A2A4J606 A0A0K8TU38 A0A212EXB5 A0A2H1VI77 A0A2H1VVY8 A0A212FJZ7 A0A212F9C7 A0A336MJ36 A0A067R2U4 A0A2H1VI74 U4URM8 R4V1J3 N6U7G6 A0A1W4WTJ1 A0A1I8NVH9 A0A1I8MK16 A0A1A9VQQ0
Pubmed
EMBL
EZ407142
ACX53704.1
RBVL01000111
RKO12019.1
MF196310
ATU07292.1
+ More
RBVL01000071 RKO12462.1 ODYU01009308 SOQ53635.1 KQ459595 KPI95817.1 AGBW02010899 OWR47538.1 KQ460473 KPJ14396.1 GDQN01000386 JAT90668.1 ODYU01000156 SOQ34451.1 GCVX01000230 JAI18000.1 BABH01039669 BABH01039670 AB196702 AB383136 BAE43410.1 BAH97092.1 EZ407252 ACX53809.1 MF196302 ATU07284.1 ODYU01001173 SOQ37016.1 KZ149964 PZC76238.1 AY231292 AAO65575.1 NWSH01000992 PCG73195.1 NWSH01001136 PCG72449.1 RBVL01000123 RKO11885.1 KZ149950 PZC76676.1 SOQ37017.1 KT443977 ANC68525.1 KT261667 MF196309 ALJ30208.1 ATU07291.1 RSAL01000004 RVE54626.1 ODYU01009376 SOQ53760.1 EZ407156 ACX53718.1 KQ460202 KPJ17177.1 JTDY01001270 KOB74335.1 JXJN01004857 SOQ34454.1 AK402595 BAM19217.1 EZ407239 ACX53796.1 MF196305 ATU07287.1 GDQN01009976 JAT81078.1 PZC76239.1 EZ407269 ACX53824.1 AK401870 BAM18492.1 KPI95934.1 OWR47537.1 ODYU01007918 SOQ51114.1 NWSH01001445 PCG71216.1 NEVH01011194 PNF31891.1 KQ459299 KPJ01910.1 KT443975 ANC68523.1 KY419208 AVC68634.1 PNF31889.1 GCVX01000231 JAI17999.1 RBVL01000055 RKO12710.1 MF196312 ATU07294.1 GCVX01000235 JAI17995.1 RSAL01000076 RVE48836.1 KQ460313 KPJ16090.1 GAIX01014897 JAA77663.1 PCG71215.1 ODYU01006397 SOQ48206.1 PNF31911.1 PCG73196.1 KK856142 PTY25006.1 OUUW01000008 SPP83812.1 MF196296 ATU07278.1 RBVL01000182 RKO11020.1 PNF31890.1 KPJ14400.1 BABH01019418 KX585301 ARO70194.1 ODYU01001950 SOQ38859.1 KT443979 ANC68527.1 AB196700 BAE43408.1 AB383144 BAH97100.1 GBHO01010986 GBRD01003379 GDHC01008731 JAG32618.1 JAG62442.1 JAQ09898.1 GCVX01000229 JAI18001.1 PCG72453.1 ODYU01012978 SOQ59553.1 SPP83813.1 KT261655 MF196311 ALJ30196.1 ATU07293.1 NWSH01002781 PCG67547.1 GCVX01000233 JAI17997.1 AGBW02011789 OWR46125.1 ODYU01002699 SOQ40539.1 ODYU01004766 SOQ44995.1 AGBW02008184 OWR54054.1 AGBW02009605 OWR50342.1 UFQS01000646 UFQT01000646 SSX05709.1 SSX26068.1 KK852744 KDR17338.1 SOQ40537.1 KB632409 ERL95168.1 KC740703 AGM32527.1 APGK01036621 KB740941 ENN77585.1
RBVL01000071 RKO12462.1 ODYU01009308 SOQ53635.1 KQ459595 KPI95817.1 AGBW02010899 OWR47538.1 KQ460473 KPJ14396.1 GDQN01000386 JAT90668.1 ODYU01000156 SOQ34451.1 GCVX01000230 JAI18000.1 BABH01039669 BABH01039670 AB196702 AB383136 BAE43410.1 BAH97092.1 EZ407252 ACX53809.1 MF196302 ATU07284.1 ODYU01001173 SOQ37016.1 KZ149964 PZC76238.1 AY231292 AAO65575.1 NWSH01000992 PCG73195.1 NWSH01001136 PCG72449.1 RBVL01000123 RKO11885.1 KZ149950 PZC76676.1 SOQ37017.1 KT443977 ANC68525.1 KT261667 MF196309 ALJ30208.1 ATU07291.1 RSAL01000004 RVE54626.1 ODYU01009376 SOQ53760.1 EZ407156 ACX53718.1 KQ460202 KPJ17177.1 JTDY01001270 KOB74335.1 JXJN01004857 SOQ34454.1 AK402595 BAM19217.1 EZ407239 ACX53796.1 MF196305 ATU07287.1 GDQN01009976 JAT81078.1 PZC76239.1 EZ407269 ACX53824.1 AK401870 BAM18492.1 KPI95934.1 OWR47537.1 ODYU01007918 SOQ51114.1 NWSH01001445 PCG71216.1 NEVH01011194 PNF31891.1 KQ459299 KPJ01910.1 KT443975 ANC68523.1 KY419208 AVC68634.1 PNF31889.1 GCVX01000231 JAI17999.1 RBVL01000055 RKO12710.1 MF196312 ATU07294.1 GCVX01000235 JAI17995.1 RSAL01000076 RVE48836.1 KQ460313 KPJ16090.1 GAIX01014897 JAA77663.1 PCG71215.1 ODYU01006397 SOQ48206.1 PNF31911.1 PCG73196.1 KK856142 PTY25006.1 OUUW01000008 SPP83812.1 MF196296 ATU07278.1 RBVL01000182 RKO11020.1 PNF31890.1 KPJ14400.1 BABH01019418 KX585301 ARO70194.1 ODYU01001950 SOQ38859.1 KT443979 ANC68527.1 AB196700 BAE43408.1 AB383144 BAH97100.1 GBHO01010986 GBRD01003379 GDHC01008731 JAG32618.1 JAG62442.1 JAQ09898.1 GCVX01000229 JAI18001.1 PCG72453.1 ODYU01012978 SOQ59553.1 SPP83813.1 KT261655 MF196311 ALJ30196.1 ATU07293.1 NWSH01002781 PCG67547.1 GCVX01000233 JAI17997.1 AGBW02011789 OWR46125.1 ODYU01002699 SOQ40539.1 ODYU01004766 SOQ44995.1 AGBW02008184 OWR54054.1 AGBW02009605 OWR50342.1 UFQS01000646 UFQT01000646 SSX05709.1 SSX26068.1 KK852744 KDR17338.1 SOQ40537.1 KB632409 ERL95168.1 KC740703 AGM32527.1 APGK01036621 KB740941 ENN77585.1
Proteomes
Interpro
SUPFAM
SSF51126
SSF51126
Gene 3D
ProteinModelPortal
D2SNL3
A0A3B0EWA9
A0A3G1KLC0
A0A3B0ET75
A0A2H1WKM4
A0A194PR79
+ More
A0A212F1C0 A0A194REN7 A0A1E1WUL1 A0A2H1V0V6 A0A0K8TVA7 H9IVP4 Q402D8 D2SNW8 A0A3G1KKW8 A0A2H1V896 A0A2W1BMT1 Q86GJ4 A0A2A4JM86 A0A2A4JKV9 A0A3B0EWC7 A0A2W1BVK0 A0A2H1V876 A0A161IWD8 A0A0P0EUL4 A0A3S2P7X1 A0A2H1WKZ8 D2SNM7 A0A0N0PDH3 A0A0L7LG13 A0A1A9YIN2 A0A1B0AWP1 A0A2H1V2G1 I4DMS7 D2SNV5 A0A3G1KL03 A0A1E1W258 A0A2W1BMF2 D2SNY3 I4DKQ2 A0A212F1D1 A0A2H1WF65 A0A2A4JHG1 A0A2J7QTH9 A0A194QA85 A0A168T0I5 A0A3G1NI93 A0A2J7QTH3 A0A0K8TU33 A0A3B0EYT6 A0A3G1KKZ9 A0A0K8TVA3 A0A3S2M186 A0A194RFL6 S4NQ33 A0A2A4JGK7 A0A2H1W5H9 A0A2J7QTJ5 A0A2A4JN36 A0A2R7WZQ2 A0A3B0JTT6 A0A3G1KKW6 A0A3B0EU73 A0A2J7QTH5 A0A194RAT9 H9JNH2 A0A1B0A461 A0A2K8GKQ0 A0A2H1VDE1 A0A161JCG7 Q402E0 C6KYM6 A0A0A9YNG7 A0A0K8TUQ8 A0A2A4JLY9 A0A2H1X2K5 A0A3B0JS36 A0A0P0EUK7 A0A2A4J606 A0A0K8TU38 A0A212EXB5 A0A2H1VI77 A0A2H1VVY8 A0A212FJZ7 A0A212F9C7 A0A336MJ36 A0A067R2U4 A0A2H1VI74 U4URM8 R4V1J3 N6U7G6 A0A1W4WTJ1 A0A1I8NVH9 A0A1I8MK16 A0A1A9VQQ0
A0A212F1C0 A0A194REN7 A0A1E1WUL1 A0A2H1V0V6 A0A0K8TVA7 H9IVP4 Q402D8 D2SNW8 A0A3G1KKW8 A0A2H1V896 A0A2W1BMT1 Q86GJ4 A0A2A4JM86 A0A2A4JKV9 A0A3B0EWC7 A0A2W1BVK0 A0A2H1V876 A0A161IWD8 A0A0P0EUL4 A0A3S2P7X1 A0A2H1WKZ8 D2SNM7 A0A0N0PDH3 A0A0L7LG13 A0A1A9YIN2 A0A1B0AWP1 A0A2H1V2G1 I4DMS7 D2SNV5 A0A3G1KL03 A0A1E1W258 A0A2W1BMF2 D2SNY3 I4DKQ2 A0A212F1D1 A0A2H1WF65 A0A2A4JHG1 A0A2J7QTH9 A0A194QA85 A0A168T0I5 A0A3G1NI93 A0A2J7QTH3 A0A0K8TU33 A0A3B0EYT6 A0A3G1KKZ9 A0A0K8TVA3 A0A3S2M186 A0A194RFL6 S4NQ33 A0A2A4JGK7 A0A2H1W5H9 A0A2J7QTJ5 A0A2A4JN36 A0A2R7WZQ2 A0A3B0JTT6 A0A3G1KKW6 A0A3B0EU73 A0A2J7QTH5 A0A194RAT9 H9JNH2 A0A1B0A461 A0A2K8GKQ0 A0A2H1VDE1 A0A161JCG7 Q402E0 C6KYM6 A0A0A9YNG7 A0A0K8TUQ8 A0A2A4JLY9 A0A2H1X2K5 A0A3B0JS36 A0A0P0EUK7 A0A2A4J606 A0A0K8TU38 A0A212EXB5 A0A2H1VI77 A0A2H1VVY8 A0A212FJZ7 A0A212F9C7 A0A336MJ36 A0A067R2U4 A0A2H1VI74 U4URM8 R4V1J3 N6U7G6 A0A1W4WTJ1 A0A1I8NVH9 A0A1I8MK16 A0A1A9VQQ0
Ontologies
Topology
SignalP
Position: 1 - 17,
Likelihood: 0.985175
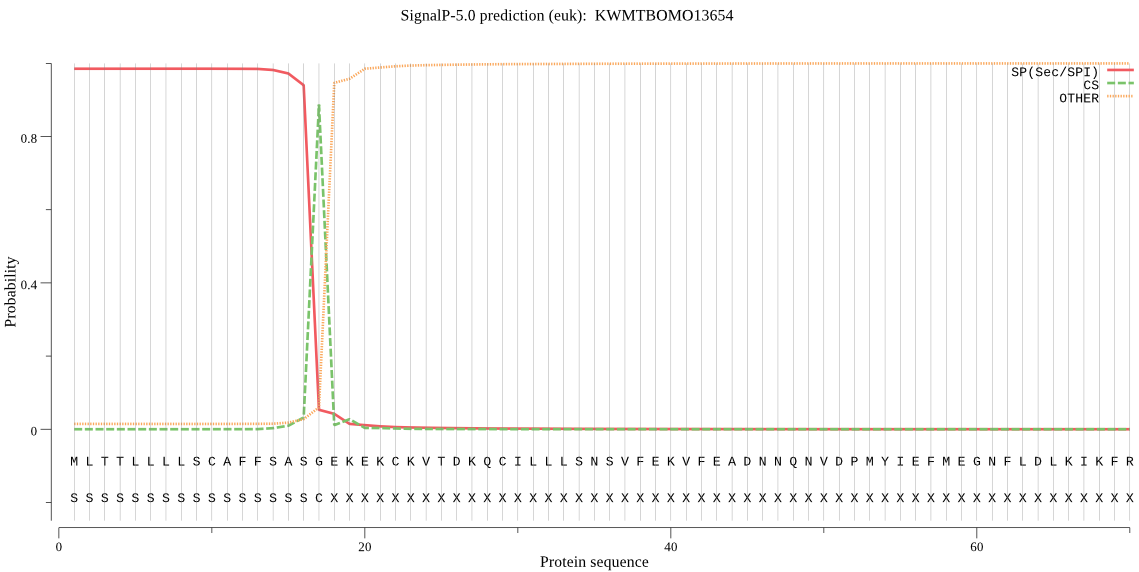
Length:
228
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01972
Exp number, first 60 AAs:
0.00046
Total prob of N-in:
0.00875
outside
1 - 228

Population Genetic Test Statistics
Pi
184.107854
Theta
136.865819
Tajima's D
0.992659
CLR
0.067185
CSRT
0.658517074146293
Interpretation
Uncertain
