Pre Gene Modal
BGIBMGA011039
Annotation
PREDICTED:_structural_maintenance_of_chromosomes_protein_3_[Amyelois_transitella]
Full name
Structural maintenance of chromosomes protein
Location in the cell
Nuclear Reliability : 2.914
Sequence
CDS
ATGCACATAAAACAGGTAATAATCCAAGGATTCAAGAGTTACCGAGAGCAAATTGTTGTAGAGCCCTTTGATAAACGGCATAATGTAGTCGTGGGACGTAATGGTTCAGGGAAGAGTAACTTTTTCCACGCCATTCAATTTGTACTCAGCGATGAATTCTCCCATCTCAGACCTGAGCAGCGGCTGGCCCTACTGCACGAGGGAACTGGCCCTAGAGTCATTTCGGCTTTTGTTGAAATTATATTTGACAACTCTGACAATAGGATACCTATTGAAAAAGATGAGATATTTTTACGACGTGTCATTGGTTCAAAGAAGGATCAGTTCTTCCTCAACAAAAAAGTCGTTCCTCGTTCAGAAGTACTAAACTTGCTTGAGAGTGCTGGGCTTTCTAATTCCAATCCATATTACATTGTGAAACAGGGAAAAATTAATCAAATGGCCATAGCTCCTGATTCTCACAGACTGAAACTTCTAAGAGAAGTAGCTGGTACAAGGGTTTACGACGAAAGGAGGGAGGAGTCTGTAGCTATACTTAAAGAGACTGTTGGAAAGGTAGAAAAAATAAATGAATTCCTCCAAACCATTGAAGAACGTTTGAAAACTTTAGAAGAGGAGAAGGAAGAATTAAAAGAGTACCAGAAATGGGACAGAGCTAGACGCGTTCTAGAATTTATTATCCATGACACTGAACACAAGGAGAACAAAAGGAAACTGGAAGAGTTGGAGAAACTGAGATCGAACAGTGGAAAAGAACAACAACATTATGCAGATTTGGTACGAGAAGCTCAAGAACACGTGAGGGAAGCTAATAGGAAGCTGAAGGAAGCGCGCAAGGACGTGGCGGCTGCAAGAGAGGAAAAGGACATCCTGTCCACCGAACAACAACAGCTGCTGAAGGAAAAAACGAAATTAGAACTTGCCATAAAGGACTTGACCGACGACGTTGACGGCGACAACAAATCGAAGGAACGCGCTGAAGCCGAATTGGAACGCTTAAGACAGCAAATATCAGAGAAAGAACGCGAGTTAGAAGAACTGAAGCCGAAATATGAGGAGATGAAGGCTCGTGAAGAGGAATGTACTAGAGCGCTCTCTCTCAACCAGCAGAAGAGACAGGAGTTGTACGCCAAGCAGGGGAGAGGCACGCAGTTCACCTCCAAGCAGGACCGGGACAGATGGATCGAGAAGGAGCTCAAATCACTGAACAAGCAATTGAAAGACAAGAAAGATCACGAGAGTAAGTTACGCGAGGATCTAAGGCGCGACGCCAACAAACTGACAGAACTGGAGAAGAGAATCGAAGAGACCACGAAAGAGATGGAGAGGCAGCGCGTGGCCATCGACGAGCACAACAAGCAGTATTACGAATGTAAGAAGAAGAAGGACCAAGAACAGAGCGCTAGGAACGAACTGTGGCGCAAAGAGACTTCATTGACACAAAATTTGTCGTCACTAAAAGACGACTTGGCAAAGGCCGATCAGGCGTTACGATCCATGGCGGGAAAGCCAATTCTGAACGGTCGAGACAGTGTCCGCAAGGTGCTGGAGACTTTCCAGGAGCGCGGAGGCGACTGGGCCAAAATAGCCACCCAGTACTACGGGCCCGTCATCGAGAACTTCACCTGCGATAAGACTATCTACACGGCCGTCGAGGTAACAGCGGGCAACAGACTGTTCCATCACATCGTAGAATCCGACACTGTAGGAACGAAGATCCTCAAAGAGATGAACAGACAGAACCTACCCGGAGAAGTCACCTTCATGCCCTTGAACCGACTGCAAGTACGAGATATGGTTTATCCCAATGACAATAACGCAATAGCGATGGTGCAAAAACTCAAATATGATCCGAAATACGCGAAAGCAATGAAGTACATATTCGGGAAGACGCTCATCTGCAGGAATCTCGAGTGCGCCACGGAACTGGGGAAGCAGTTCCATCTGGATTGTGTCACTTTAGAAGGAGATCAGGTATCGTCGAAGGGCTCTCTGACGGGTGGTTACTTTAACCAGTCGCGCTCTCGTCTGGAAATGCAGAAGACCCGCTCCGAGCTGATGGAGCAGATCACGACCCTCGACCTGGAGCTCAGCACGCTGCGGCAGGAACTCAACAAGACTGAGGCGAGCATCAACAGCATCGTGTCCGAGATGCAGAGGACTGAGACTAAACAAGGGAAAGCAAAGGATATTTTCGACAAAGTCAAAGCAGACATTCGTCTGATGAAAGAAGAGTTAGCTTCCATAGAAAGGTTCAGAGGACCTAAGGAGAGATCTTTAGCGCAGTGCAGGTCCAGCCTGGAAGCCATGCAGGCTACCAAAGAAGGATTGGAATCTGAACTTCATCAGGAGTTGATGGAGCAACTATCGACCGCGGACCAAGGTAAAGTGGACGAACTGAACGACGCGATTCGCCGCCTCACCCTCGAGAACAAGGAAGCGTTCAGCGCCAGAATGAATCTCGAAGCCACAAAGAACAAACTAGAGAATCTGCTGACCAACAATTTGATTCGGTTCGTTCATCGTTCGTTCGATAGTTTGTGTAAAGTTACCGAATTTAAACCTCTAAAAATAATATATTAG
Protein
MHIKQVIIQGFKSYREQIVVEPFDKRHNVVVGRNGSGKSNFFHAIQFVLSDEFSHLRPEQRLALLHEGTGPRVISAFVEIIFDNSDNRIPIEKDEIFLRRVIGSKKDQFFLNKKVVPRSEVLNLLESAGLSNSNPYYIVKQGKINQMAIAPDSHRLKLLREVAGTRVYDERREESVAILKETVGKVEKINEFLQTIEERLKTLEEEKEELKEYQKWDRARRVLEFIIHDTEHKENKRKLEELEKLRSNSGKEQQHYADLVREAQEHVREANRKLKEARKDVAAAREEKDILSTEQQQLLKEKTKLELAIKDLTDDVDGDNKSKERAEAELERLRQQISEKERELEELKPKYEEMKAREEECTRALSLNQQKRQELYAKQGRGTQFTSKQDRDRWIEKELKSLNKQLKDKKDHESKLREDLRRDANKLTELEKRIEETTKEMERQRVAIDEHNKQYYECKKKKDQEQSARNELWRKETSLTQNLSSLKDDLAKADQALRSMAGKPILNGRDSVRKVLETFQERGGDWAKIATQYYGPVIENFTCDKTIYTAVEVTAGNRLFHHIVESDTVGTKILKEMNRQNLPGEVTFMPLNRLQVRDMVYPNDNNAIAMVQKLKYDPKYAKAMKYIFGKTLICRNLECATELGKQFHLDCVTLEGDQVSSKGSLTGGYFNQSRSRLEMQKTRSELMEQITTLDLELSTLRQELNKTEASINSIVSEMQRTETKQGKAKDIFDKVKADIRLMKEELASIERFRGPKERSLAQCRSSLEAMQATKEGLESELHQELMEQLSTADQGKVDELNDAIRRLTLENKEAFSARMNLEATKNKLENLLTNNLIRFVHRSFDSLCKVTEFKPLKIIY
Summary
Similarity
Belongs to the SMC family.
Feature
chain Structural maintenance of chromosomes protein
Uniprot
H9JND6
A0A2A4JG49
A0A2W1BPW1
A0A212EKQ6
A0A2H1V902
A0A1Y1M794
+ More
D6X4W0 A0A2J7PN85 A0A1Q3G1Y3 Q174C2 A0A336KXB2 A0A084VQJ7 A0A182NIF4 A0A2M3Z3F4 A0A182J5N9 A0A182MHM9 A0A182QYS1 A0A2M4AEF9 A0A182YLV7 A0A182JZ96 A0A182PET4 A0A182WC99 A0A182FK52 A0A182R5J8 W5JIT8 A0A1B0CNW5 A0A182V9Z7 A0A182ULJ0 A0A182WTU4 Q8I952 A0A182HI41 A0A182FJH0 A0A067R1S0 A0A0C9RMH9 A0A291S6Y0 A0A069DY14 A0A224XGP7 A0A195FNQ0 A0A151IVP0 A0A151XDI3 F4WJI3 A0A1B6DW45 E2BWB1 A0A1I8NHL7 A0A1A9W229 A0A0A9X6G6 A0A0K8SN12 A0A2A3ECT5 A0A088AHL5 A0A1B6KHR2 A0A1I8PFU5 A0A1B0G8C8 A0A1A9VBM6 A0A1A9Z5U2 A0A1A9XXS4 A0A1B0APR8 A0A1I8PG28 E2AIG6 A0A0J7NYK5 A0A0M9A885 W8C2R6 E9J3C0 A0A0A1XI14 A0A0K8VN34 A0A034WPC9 A0A034WQZ0 H0RNH8 K7J192 B4PXU7 T1HYT8 B3MSI1 B4NDL4 Q9VXE9 Q24098 B3NVJ1 A0A0L7RIJ8 A0A1W4VJZ6 Q494K8 A0A0M4EVH5 B4L846 Q29HM9 N6TVH3 A0A3B0JTJ2 J9JRW8 B4JWY8 A0A0K2ULS3 A0A2S2QFV7 E0VYD1 A0A2H8TE27 A0A195D1H2 B4M2X6 J9JYS1 A0A0P5QNA3 A0A0P5PNC9 A0A0N8C2B2 A0A026WYQ5 A0A0P5PJB9 A0A0P5YXW5 A0A2S2R9T3
D6X4W0 A0A2J7PN85 A0A1Q3G1Y3 Q174C2 A0A336KXB2 A0A084VQJ7 A0A182NIF4 A0A2M3Z3F4 A0A182J5N9 A0A182MHM9 A0A182QYS1 A0A2M4AEF9 A0A182YLV7 A0A182JZ96 A0A182PET4 A0A182WC99 A0A182FK52 A0A182R5J8 W5JIT8 A0A1B0CNW5 A0A182V9Z7 A0A182ULJ0 A0A182WTU4 Q8I952 A0A182HI41 A0A182FJH0 A0A067R1S0 A0A0C9RMH9 A0A291S6Y0 A0A069DY14 A0A224XGP7 A0A195FNQ0 A0A151IVP0 A0A151XDI3 F4WJI3 A0A1B6DW45 E2BWB1 A0A1I8NHL7 A0A1A9W229 A0A0A9X6G6 A0A0K8SN12 A0A2A3ECT5 A0A088AHL5 A0A1B6KHR2 A0A1I8PFU5 A0A1B0G8C8 A0A1A9VBM6 A0A1A9Z5U2 A0A1A9XXS4 A0A1B0APR8 A0A1I8PG28 E2AIG6 A0A0J7NYK5 A0A0M9A885 W8C2R6 E9J3C0 A0A0A1XI14 A0A0K8VN34 A0A034WPC9 A0A034WQZ0 H0RNH8 K7J192 B4PXU7 T1HYT8 B3MSI1 B4NDL4 Q9VXE9 Q24098 B3NVJ1 A0A0L7RIJ8 A0A1W4VJZ6 Q494K8 A0A0M4EVH5 B4L846 Q29HM9 N6TVH3 A0A3B0JTJ2 J9JRW8 B4JWY8 A0A0K2ULS3 A0A2S2QFV7 E0VYD1 A0A2H8TE27 A0A195D1H2 B4M2X6 J9JYS1 A0A0P5QNA3 A0A0P5PNC9 A0A0N8C2B2 A0A026WYQ5 A0A0P5PJB9 A0A0P5YXW5 A0A2S2R9T3
Pubmed
19121390
28756777
22118469
28004739
18362917
19820115
+ More
17510324 24438588 25244985 20920257 23761445 12364791 14660695 14747013 17210077 24845553 28992199 26334808 21719571 20798317 25315136 25401762 26823975 24495485 21282665 25830018 25348373 20075255 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 8849894 15632085 23537049 20566863 24508170
17510324 24438588 25244985 20920257 23761445 12364791 14660695 14747013 17210077 24845553 28992199 26334808 21719571 20798317 25315136 25401762 26823975 24495485 21282665 25830018 25348373 20075255 17994087 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 8849894 15632085 23537049 20566863 24508170
EMBL
BABH01019345
NWSH01001703
PCG70393.1
KZ149988
PZC75644.1
AGBW02014225
+ More
OWR42055.1 ODYU01001305 SOQ37330.1 GEZM01038226 JAV81709.1 KQ971381 EEZ97230.1 NEVH01023957 PNF17803.1 GFDL01001218 JAV33827.1 CH477414 EAT41403.1 UFQS01001145 UFQT01001092 UFQT01001145 SSX09245.1 SSX28872.1 ATLV01015219 KE525003 KFB40241.1 GGFM01002296 MBW23047.1 AXCM01000417 AXCN02000886 GGFK01005845 MBW39166.1 ADMH02001377 ETN62709.1 AJWK01021040 AJ535205 AAAB01008960 CAD59405.1 EAA11190.3 APCN01005875 KK852811 KDR15942.1 GBYB01014617 JAG84384.1 MF433018 ATL75369.1 GBGD01000168 JAC88721.1 GFTR01008664 JAW07762.1 KQ981387 KYN42041.1 KQ980910 KYN11475.1 KQ982294 KYQ58409.1 GL888183 EGI65639.1 GEDC01007431 JAS29867.1 GL451103 EFN80015.1 GBHO01030949 GBHO01006689 GDHC01020929 GDHC01000271 JAG12655.1 JAG36915.1 JAP97699.1 JAQ18358.1 GBRD01011092 JAG54732.1 KZ288282 PBC29520.1 GEBQ01028996 JAT10981.1 CCAG010000239 JXJN01001549 GL439791 EFN66756.1 LBMM01000789 KMQ97485.1 KQ435727 KOX77869.1 GAMC01003042 JAC03514.1 GL768061 EFZ12663.1 GBXI01003732 JAD10560.1 GDHF01012022 JAI40292.1 GAKP01002785 JAC56167.1 GAKP01002784 JAC56168.1 BT132900 AEV53908.1 CM000162 EDX01933.1 ACPB03022213 CH902622 EDV34736.2 CH964239 EDW81836.1 AE014298 AAF48625.2 U30492 AAC47078.1 CH954180 EDV46383.1 KQ414584 KOC70556.1 BT023768 AAZ41776.1 CP012528 ALC49272.1 CH933814 EDW05621.1 CH379064 EAL31729.2 APGK01053201 KB741222 KB631665 ENN72401.1 ERL85147.1 OUUW01000003 SPP78800.1 ABLF02028811 CH916376 EDV95264.1 HACA01021466 CDW38827.1 GGMS01007413 MBY76616.1 DS235845 EEB18387.1 GFXV01000520 MBW12325.1 KQ976973 KYN06753.1 CH940651 EDW65151.2 ABLF02022093 GDIQ01121764 JAL29962.1 GDIQ01125970 JAL25756.1 GDIQ01121763 JAL29963.1 KK107063 EZA60968.1 GDIQ01127562 JAL24164.1 GDIP01053038 JAM50677.1 GGMS01017520 MBY86723.1
OWR42055.1 ODYU01001305 SOQ37330.1 GEZM01038226 JAV81709.1 KQ971381 EEZ97230.1 NEVH01023957 PNF17803.1 GFDL01001218 JAV33827.1 CH477414 EAT41403.1 UFQS01001145 UFQT01001092 UFQT01001145 SSX09245.1 SSX28872.1 ATLV01015219 KE525003 KFB40241.1 GGFM01002296 MBW23047.1 AXCM01000417 AXCN02000886 GGFK01005845 MBW39166.1 ADMH02001377 ETN62709.1 AJWK01021040 AJ535205 AAAB01008960 CAD59405.1 EAA11190.3 APCN01005875 KK852811 KDR15942.1 GBYB01014617 JAG84384.1 MF433018 ATL75369.1 GBGD01000168 JAC88721.1 GFTR01008664 JAW07762.1 KQ981387 KYN42041.1 KQ980910 KYN11475.1 KQ982294 KYQ58409.1 GL888183 EGI65639.1 GEDC01007431 JAS29867.1 GL451103 EFN80015.1 GBHO01030949 GBHO01006689 GDHC01020929 GDHC01000271 JAG12655.1 JAG36915.1 JAP97699.1 JAQ18358.1 GBRD01011092 JAG54732.1 KZ288282 PBC29520.1 GEBQ01028996 JAT10981.1 CCAG010000239 JXJN01001549 GL439791 EFN66756.1 LBMM01000789 KMQ97485.1 KQ435727 KOX77869.1 GAMC01003042 JAC03514.1 GL768061 EFZ12663.1 GBXI01003732 JAD10560.1 GDHF01012022 JAI40292.1 GAKP01002785 JAC56167.1 GAKP01002784 JAC56168.1 BT132900 AEV53908.1 CM000162 EDX01933.1 ACPB03022213 CH902622 EDV34736.2 CH964239 EDW81836.1 AE014298 AAF48625.2 U30492 AAC47078.1 CH954180 EDV46383.1 KQ414584 KOC70556.1 BT023768 AAZ41776.1 CP012528 ALC49272.1 CH933814 EDW05621.1 CH379064 EAL31729.2 APGK01053201 KB741222 KB631665 ENN72401.1 ERL85147.1 OUUW01000003 SPP78800.1 ABLF02028811 CH916376 EDV95264.1 HACA01021466 CDW38827.1 GGMS01007413 MBY76616.1 DS235845 EEB18387.1 GFXV01000520 MBW12325.1 KQ976973 KYN06753.1 CH940651 EDW65151.2 ABLF02022093 GDIQ01121764 JAL29962.1 GDIQ01125970 JAL25756.1 GDIQ01121763 JAL29963.1 KK107063 EZA60968.1 GDIQ01127562 JAL24164.1 GDIP01053038 JAM50677.1 GGMS01017520 MBY86723.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000007266
UP000235965
UP000008820
+ More
UP000030765 UP000075884 UP000075880 UP000075883 UP000075886 UP000076408 UP000075881 UP000075885 UP000075920 UP000069272 UP000075900 UP000000673 UP000092461 UP000075903 UP000075902 UP000076407 UP000007062 UP000075840 UP000027135 UP000078541 UP000078492 UP000075809 UP000007755 UP000008237 UP000095301 UP000091820 UP000242457 UP000005203 UP000095300 UP000092444 UP000078200 UP000092445 UP000092443 UP000092460 UP000000311 UP000036403 UP000053105 UP000002358 UP000002282 UP000015103 UP000007801 UP000007798 UP000000803 UP000008711 UP000053825 UP000192221 UP000092553 UP000009192 UP000001819 UP000019118 UP000030742 UP000268350 UP000007819 UP000001070 UP000009046 UP000078542 UP000008792 UP000053097
UP000030765 UP000075884 UP000075880 UP000075883 UP000075886 UP000076408 UP000075881 UP000075885 UP000075920 UP000069272 UP000075900 UP000000673 UP000092461 UP000075903 UP000075902 UP000076407 UP000007062 UP000075840 UP000027135 UP000078541 UP000078492 UP000075809 UP000007755 UP000008237 UP000095301 UP000091820 UP000242457 UP000005203 UP000095300 UP000092444 UP000078200 UP000092445 UP000092443 UP000092460 UP000000311 UP000036403 UP000053105 UP000002358 UP000002282 UP000015103 UP000007801 UP000007798 UP000000803 UP000008711 UP000053825 UP000192221 UP000092553 UP000009192 UP000001819 UP000019118 UP000030742 UP000268350 UP000007819 UP000001070 UP000009046 UP000078542 UP000008792 UP000053097
Interpro
ProteinModelPortal
H9JND6
A0A2A4JG49
A0A2W1BPW1
A0A212EKQ6
A0A2H1V902
A0A1Y1M794
+ More
D6X4W0 A0A2J7PN85 A0A1Q3G1Y3 Q174C2 A0A336KXB2 A0A084VQJ7 A0A182NIF4 A0A2M3Z3F4 A0A182J5N9 A0A182MHM9 A0A182QYS1 A0A2M4AEF9 A0A182YLV7 A0A182JZ96 A0A182PET4 A0A182WC99 A0A182FK52 A0A182R5J8 W5JIT8 A0A1B0CNW5 A0A182V9Z7 A0A182ULJ0 A0A182WTU4 Q8I952 A0A182HI41 A0A182FJH0 A0A067R1S0 A0A0C9RMH9 A0A291S6Y0 A0A069DY14 A0A224XGP7 A0A195FNQ0 A0A151IVP0 A0A151XDI3 F4WJI3 A0A1B6DW45 E2BWB1 A0A1I8NHL7 A0A1A9W229 A0A0A9X6G6 A0A0K8SN12 A0A2A3ECT5 A0A088AHL5 A0A1B6KHR2 A0A1I8PFU5 A0A1B0G8C8 A0A1A9VBM6 A0A1A9Z5U2 A0A1A9XXS4 A0A1B0APR8 A0A1I8PG28 E2AIG6 A0A0J7NYK5 A0A0M9A885 W8C2R6 E9J3C0 A0A0A1XI14 A0A0K8VN34 A0A034WPC9 A0A034WQZ0 H0RNH8 K7J192 B4PXU7 T1HYT8 B3MSI1 B4NDL4 Q9VXE9 Q24098 B3NVJ1 A0A0L7RIJ8 A0A1W4VJZ6 Q494K8 A0A0M4EVH5 B4L846 Q29HM9 N6TVH3 A0A3B0JTJ2 J9JRW8 B4JWY8 A0A0K2ULS3 A0A2S2QFV7 E0VYD1 A0A2H8TE27 A0A195D1H2 B4M2X6 J9JYS1 A0A0P5QNA3 A0A0P5PNC9 A0A0N8C2B2 A0A026WYQ5 A0A0P5PJB9 A0A0P5YXW5 A0A2S2R9T3
D6X4W0 A0A2J7PN85 A0A1Q3G1Y3 Q174C2 A0A336KXB2 A0A084VQJ7 A0A182NIF4 A0A2M3Z3F4 A0A182J5N9 A0A182MHM9 A0A182QYS1 A0A2M4AEF9 A0A182YLV7 A0A182JZ96 A0A182PET4 A0A182WC99 A0A182FK52 A0A182R5J8 W5JIT8 A0A1B0CNW5 A0A182V9Z7 A0A182ULJ0 A0A182WTU4 Q8I952 A0A182HI41 A0A182FJH0 A0A067R1S0 A0A0C9RMH9 A0A291S6Y0 A0A069DY14 A0A224XGP7 A0A195FNQ0 A0A151IVP0 A0A151XDI3 F4WJI3 A0A1B6DW45 E2BWB1 A0A1I8NHL7 A0A1A9W229 A0A0A9X6G6 A0A0K8SN12 A0A2A3ECT5 A0A088AHL5 A0A1B6KHR2 A0A1I8PFU5 A0A1B0G8C8 A0A1A9VBM6 A0A1A9Z5U2 A0A1A9XXS4 A0A1B0APR8 A0A1I8PG28 E2AIG6 A0A0J7NYK5 A0A0M9A885 W8C2R6 E9J3C0 A0A0A1XI14 A0A0K8VN34 A0A034WPC9 A0A034WQZ0 H0RNH8 K7J192 B4PXU7 T1HYT8 B3MSI1 B4NDL4 Q9VXE9 Q24098 B3NVJ1 A0A0L7RIJ8 A0A1W4VJZ6 Q494K8 A0A0M4EVH5 B4L846 Q29HM9 N6TVH3 A0A3B0JTJ2 J9JRW8 B4JWY8 A0A0K2ULS3 A0A2S2QFV7 E0VYD1 A0A2H8TE27 A0A195D1H2 B4M2X6 J9JYS1 A0A0P5QNA3 A0A0P5PNC9 A0A0N8C2B2 A0A026WYQ5 A0A0P5PJB9 A0A0P5YXW5 A0A2S2R9T3
PDB
2WD5
E-value=3.0295e-61,
Score=599
Ontologies
GO
Topology
Subcellular location
Nucleus
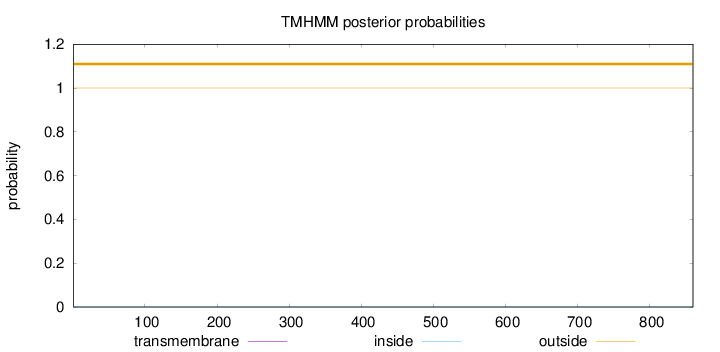
Length:
860
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00153
Exp number, first 60 AAs:
0.00057
Total prob of N-in:
0.00012
outside
1 - 860
Population Genetic Test Statistics
Pi
229.851665
Theta
176.767483
Tajima's D
1.066433
CLR
0.348423
CSRT
0.676766161691915
Interpretation
Uncertain
