Gene
KWMTBOMO13643
Pre Gene Modal
BGIBMGA011072
Annotation
PREDICTED:_lipase_3-like_[Papilio_polytes]
Full name
Lipase
Location in the cell
Extracellular Reliability : 1.429 PlasmaMembrane Reliability : 1.477
Sequence
CDS
ATGTACCGTGTTTCGCTGTTCGTTCTATTATTGCCTTTAGTAACTGCCTTCAATATATTCAGTGTTTGGAACACGGTTATCAATGGCATTACAAGTTTTTTCGGTTACGATTCGCATGTAGACGCGCGATTGAATATAAGCGAACTCGCCGCAAAATACGGATATCCGTTCGAAGAGCACCGGGTGAATACAGAGGACGGTTATATTCTGAATTTACATCGAATAAAACACGGGAGAAGTGACGCTGTTCAGAGGAGGTCCGTTGTATTTTTGATGCACGGAATTTTAGACAGCTCAGACGGTTGGGTATTGCAAGGACCGGATTCAGCGTTGGCCTATATCCTAGCGGATAGCGGCTTCGATGTATGGATGGGAAACGCCAGAGGCAACAAACACTCCTTGAAGCACGCGTCCTTAAAACCCGACGATCAACGCTTTTGGGATTTCACTTGGGAAGAGATTGGCCTTTACGATTTACCGGCGTCTATCGACTACATACTAAACGCGACGGCTGAAACCGATCTATACTACGTCGGATACTCTCAAGGTACCACGTGTTATTACGTCATGACCTCAATGAAGCCCGAATACAATAACAAAATCAAGATGATGTTCTCGTTGGCTCCGGTCGCTTGGCTGGAGCACGCCAAGAGTCCTTTGGTACAAATATTTTCGCCCGCAAACGAAATACTCGGGTACGCTTTGGCGTCGCTAAACGCACACGTGCCGAGTGCGGACTCGCTCAACAAGCTATCGATGTTAGTCTGCGTGCTGTCGCCGGTGCGATGCGACAATTTCTTGTCCAACACTTTAGGTTACAGTTCTAATGTAGACGAAGAACTGATTCCAGTTATATTGGGACACGCGCCTACAGGGTGTTCGACGTTGCAGTTCGTGCACTACGGACAGTTGGCGGCGTCGGGTAGGTTCGGCAGATACGACCACGGCAGGGAGCGGAACTTGAAGGTCTACGGGCAGGAAACGGCTCCGGACTACGCATGCGCCAACATCAAGTCCCCGGTCGTGTTGTTCCACAGCGGAGAGGATTGGTTCTCCGATAAAATAGACGTCGAGAGACTAAAGGATTGTCTGCCGAACGTTCGAGAAACCGTTTACATTGAAGACCTGAGTCACATCGATTTTGTTTACGCGAAAATTAGTAGGCACGCCGTCTACGACAAAATTGTATCGATCATAGACAACTTCGAATTTGAAAATGGTTTAAAGATAACGAGTAAATCGATGCGATATAATTTTTTTACTAAATAA
Protein
MYRVSLFVLLLPLVTAFNIFSVWNTVINGITSFFGYDSHVDARLNISELAAKYGYPFEEHRVNTEDGYILNLHRIKHGRSDAVQRRSVVFLMHGILDSSDGWVLQGPDSALAYILADSGFDVWMGNARGNKHSLKHASLKPDDQRFWDFTWEEIGLYDLPASIDYILNATAETDLYYVGYSQGTTCYYVMTSMKPEYNNKIKMMFSLAPVAWLEHAKSPLVQIFSPANEILGYALASLNAHVPSADSLNKLSMLVCVLSPVRCDNFLSNTLGYSSNVDEELIPVILGHAPTGCSTLQFVHYGQLAASGRFGRYDHGRERNLKVYGQETAPDYACANIKSPVVLFHSGEDWFSDKIDVERLKDCLPNVRETVYIEDLSHIDFVYAKISRHAVYDKIVSIIDNFEFENGLKITSKSMRYNFFTK
Summary
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Feature
chain Lipase
Uniprot
H9JNG9
A0A194PS88
A0A194R9R8
A0A2W1BL70
D2A0X3
A0A194R3I6
+ More
A0A2W1BS78 A0A067RCF7 A0A2J7QW32 A0A0T6BEX7 A0A194PMH9 A0A084W6W2 A0A194R2Y3 W5JF51 D2A0X6 A0A182JMM3 A0A194PL00 A0A182RCW3 A0A182KCP4 A0A212EKP9 A0A182FR70 A0A182H3N2 A0A182TPG6 A0A2A4IVE2 A0A2H1WUD0 A0A182V7K7 A0A067R0F9 A0A182XID0 A0A182HTB5 A0A182LFG8 Q7QBX7 A0A182Y799 A0A2H1WJP8 A0A232EX80 A0A232F2D1 B0W6G8 A0A067QLF8 A0A2H1VT20 A0A2J7PUL3 A0A139WJS1 A0A212FB51 A0A2A4K633 A0A182PA26 A0A1Y1KBU1 A0A1L8DPS2 A0A1L8DQ28 K7J8S8 A0A182MKD5 A0A0J9TKN6 A0A1L8DPY1 A0A1L8DPM5 A0A0C9RRQ5 A0A2A4JCC5 A0A3F2YX72 A0A034VW56 A0A0Q5WBH0 A0A194QUC1 A0A2W1B637 B4HWR7 A0A0J9R159 A0A1W7R7P1 Q16MD1 A0A182VQ94 A0A1A9UTK2 A0A0L7LE27 A0A0R1DRH5 B3N5D9 M9MRM3 A0A0L0C6A4 A0A194PXA6 B0W003 A0A226E1Q5 W8C916 B4P0Q9 A0A2H1X0E5 A0A2H1VWQ8 Q16F27 Q9VKT9 A0A2J7PY44 A0A1B0B8Y7 A0A194QNR0 A0A194PWK6 M9PCU5 A0A1W4W6G4 A0A2H1WZN4 H9J3W1 A0A0K8W418 A0A1S4FVZ7 A0A1D2MRG2 Q17BM4 V5GWI8 A0A067RHQ8 A0A1W4UT82 A0A2A4K6W6 A0A1Y1MRZ1 A0A1A9XQ71 A0A1B2M4Y7 A0A0L7LCZ5 A0A1W4UT02 A0A1B0CSN9
A0A2W1BS78 A0A067RCF7 A0A2J7QW32 A0A0T6BEX7 A0A194PMH9 A0A084W6W2 A0A194R2Y3 W5JF51 D2A0X6 A0A182JMM3 A0A194PL00 A0A182RCW3 A0A182KCP4 A0A212EKP9 A0A182FR70 A0A182H3N2 A0A182TPG6 A0A2A4IVE2 A0A2H1WUD0 A0A182V7K7 A0A067R0F9 A0A182XID0 A0A182HTB5 A0A182LFG8 Q7QBX7 A0A182Y799 A0A2H1WJP8 A0A232EX80 A0A232F2D1 B0W6G8 A0A067QLF8 A0A2H1VT20 A0A2J7PUL3 A0A139WJS1 A0A212FB51 A0A2A4K633 A0A182PA26 A0A1Y1KBU1 A0A1L8DPS2 A0A1L8DQ28 K7J8S8 A0A182MKD5 A0A0J9TKN6 A0A1L8DPY1 A0A1L8DPM5 A0A0C9RRQ5 A0A2A4JCC5 A0A3F2YX72 A0A034VW56 A0A0Q5WBH0 A0A194QUC1 A0A2W1B637 B4HWR7 A0A0J9R159 A0A1W7R7P1 Q16MD1 A0A182VQ94 A0A1A9UTK2 A0A0L7LE27 A0A0R1DRH5 B3N5D9 M9MRM3 A0A0L0C6A4 A0A194PXA6 B0W003 A0A226E1Q5 W8C916 B4P0Q9 A0A2H1X0E5 A0A2H1VWQ8 Q16F27 Q9VKT9 A0A2J7PY44 A0A1B0B8Y7 A0A194QNR0 A0A194PWK6 M9PCU5 A0A1W4W6G4 A0A2H1WZN4 H9J3W1 A0A0K8W418 A0A1S4FVZ7 A0A1D2MRG2 Q17BM4 V5GWI8 A0A067RHQ8 A0A1W4UT82 A0A2A4K6W6 A0A1Y1MRZ1 A0A1A9XQ71 A0A1B2M4Y7 A0A0L7LCZ5 A0A1W4UT02 A0A1B0CSN9
Pubmed
19121390
26354079
28756777
18362917
19820115
24845553
+ More
24438588 20920257 23761445 22118469 26483478 20966253 12364791 14747013 17210077 25244985 28648823 28004739 20075255 22936249 25348373 17994087 17510324 26227816 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26108605 24495485 26109357 26109356 27289101
24438588 20920257 23761445 22118469 26483478 20966253 12364791 14747013 17210077 25244985 28648823 28004739 20075255 22936249 25348373 17994087 17510324 26227816 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26108605 24495485 26109357 26109356 27289101
EMBL
BABH01019345
KQ459595
KPI95828.1
KQ460473
KPJ14387.1
KZ149988
+ More
PZC75638.1 KQ971338 EFA01616.1 KQ460949 KPJ10406.1 KZ149971 PZC76037.1 KK852794 KDR16428.1 NEVH01009768 PNF32784.1 LJIG01001091 KRT85849.1 KQ459598 KPI94527.1 ATLV01020990 KE525311 KFB45956.1 KQ460855 KPJ11879.1 ADMH02001587 ETN61958.1 EFA01613.1 KQ459601 KPI94007.1 AGBW02014225 OWR42056.1 JXUM01107700 KQ565165 KXJ71265.1 NWSH01005801 PCG63937.1 ODYU01011128 SOQ56675.1 KK852827 KDR15388.1 APCN01000436 AAAB01008859 EAA08216.4 ODYU01009113 SOQ53300.1 NNAY01001790 OXU22918.1 NNAY01001167 OXU24914.1 DS231848 EDS36642.1 KK853197 KDR09957.1 ODYU01004246 SOQ43916.1 NEVH01021196 PNF20013.1 KYB28091.1 AGBW02009374 OWR50972.1 NWSH01000119 PCG79374.1 GEZM01086956 JAV58943.1 GFDF01005645 JAV08439.1 GFDF01005644 JAV08440.1 AXCM01001786 CM002910 KMY89575.1 GFDF01005647 JAV08437.1 GFDF01005646 JAV08438.1 GBYB01010056 JAG79823.1 NWSH01002126 PCG69082.1 AXCN02000051 GAKP01013224 JAC45728.1 CH954177 KQS70596.1 KQ461190 KPJ07151.1 KZ150291 PZC71579.1 CH480818 EDW52462.1 KMY89574.1 GEHC01000530 JAV47115.1 CH477866 EAT35487.1 JTDY01001496 KOB73727.1 CM000157 KRJ97507.1 EDV59018.1 AE014134 ADV37035.1 JRES01000842 KNC27786.1 KQ459590 KPI97394.1 DS231816 EDS37575.1 LNIX01000009 OXA50396.1 GAMC01007031 JAB99524.1 EDW87954.1 ODYU01012460 SOQ58781.1 ODYU01004598 SOQ44644.1 CH478517 EAT32843.1 AY089586 AAF52971.1 AAL90324.1 NEVH01020850 PNF21255.1 JXJN01010106 KPJ07152.1 KPI97393.1 AGB92891.1 ODYU01012265 SOQ58477.1 BABH01028866 GDHF01006507 JAI45807.1 LJIJ01000666 ODM95481.1 CH477320 EAT43642.1 GALX01003858 JAB64608.1 KK852673 KDR18742.1 NWSH01000114 PCG79412.1 GEZM01023396 JAV88411.1 KX447604 AOA60271.1 JTDY01001637 KOB73274.1 AJWK01026332
PZC75638.1 KQ971338 EFA01616.1 KQ460949 KPJ10406.1 KZ149971 PZC76037.1 KK852794 KDR16428.1 NEVH01009768 PNF32784.1 LJIG01001091 KRT85849.1 KQ459598 KPI94527.1 ATLV01020990 KE525311 KFB45956.1 KQ460855 KPJ11879.1 ADMH02001587 ETN61958.1 EFA01613.1 KQ459601 KPI94007.1 AGBW02014225 OWR42056.1 JXUM01107700 KQ565165 KXJ71265.1 NWSH01005801 PCG63937.1 ODYU01011128 SOQ56675.1 KK852827 KDR15388.1 APCN01000436 AAAB01008859 EAA08216.4 ODYU01009113 SOQ53300.1 NNAY01001790 OXU22918.1 NNAY01001167 OXU24914.1 DS231848 EDS36642.1 KK853197 KDR09957.1 ODYU01004246 SOQ43916.1 NEVH01021196 PNF20013.1 KYB28091.1 AGBW02009374 OWR50972.1 NWSH01000119 PCG79374.1 GEZM01086956 JAV58943.1 GFDF01005645 JAV08439.1 GFDF01005644 JAV08440.1 AXCM01001786 CM002910 KMY89575.1 GFDF01005647 JAV08437.1 GFDF01005646 JAV08438.1 GBYB01010056 JAG79823.1 NWSH01002126 PCG69082.1 AXCN02000051 GAKP01013224 JAC45728.1 CH954177 KQS70596.1 KQ461190 KPJ07151.1 KZ150291 PZC71579.1 CH480818 EDW52462.1 KMY89574.1 GEHC01000530 JAV47115.1 CH477866 EAT35487.1 JTDY01001496 KOB73727.1 CM000157 KRJ97507.1 EDV59018.1 AE014134 ADV37035.1 JRES01000842 KNC27786.1 KQ459590 KPI97394.1 DS231816 EDS37575.1 LNIX01000009 OXA50396.1 GAMC01007031 JAB99524.1 EDW87954.1 ODYU01012460 SOQ58781.1 ODYU01004598 SOQ44644.1 CH478517 EAT32843.1 AY089586 AAF52971.1 AAL90324.1 NEVH01020850 PNF21255.1 JXJN01010106 KPJ07152.1 KPI97393.1 AGB92891.1 ODYU01012265 SOQ58477.1 BABH01028866 GDHF01006507 JAI45807.1 LJIJ01000666 ODM95481.1 CH477320 EAT43642.1 GALX01003858 JAB64608.1 KK852673 KDR18742.1 NWSH01000114 PCG79412.1 GEZM01023396 JAV88411.1 KX447604 AOA60271.1 JTDY01001637 KOB73274.1 AJWK01026332
Proteomes
UP000005204
UP000053268
UP000053240
UP000007266
UP000027135
UP000235965
+ More
UP000030765 UP000000673 UP000075880 UP000075900 UP000075881 UP000007151 UP000069272 UP000069940 UP000249989 UP000075902 UP000218220 UP000075903 UP000076407 UP000075840 UP000075882 UP000007062 UP000076408 UP000215335 UP000002320 UP000075885 UP000002358 UP000075883 UP000075886 UP000008711 UP000001292 UP000008820 UP000075920 UP000078200 UP000037510 UP000002282 UP000000803 UP000037069 UP000198287 UP000092460 UP000192223 UP000094527 UP000192221 UP000092443 UP000092461
UP000030765 UP000000673 UP000075880 UP000075900 UP000075881 UP000007151 UP000069272 UP000069940 UP000249989 UP000075902 UP000218220 UP000075903 UP000076407 UP000075840 UP000075882 UP000007062 UP000076408 UP000215335 UP000002320 UP000075885 UP000002358 UP000075883 UP000075886 UP000008711 UP000001292 UP000008820 UP000075920 UP000078200 UP000037510 UP000002282 UP000000803 UP000037069 UP000198287 UP000092460 UP000192223 UP000094527 UP000192221 UP000092443 UP000092461
Interpro
Gene 3D
ProteinModelPortal
H9JNG9
A0A194PS88
A0A194R9R8
A0A2W1BL70
D2A0X3
A0A194R3I6
+ More
A0A2W1BS78 A0A067RCF7 A0A2J7QW32 A0A0T6BEX7 A0A194PMH9 A0A084W6W2 A0A194R2Y3 W5JF51 D2A0X6 A0A182JMM3 A0A194PL00 A0A182RCW3 A0A182KCP4 A0A212EKP9 A0A182FR70 A0A182H3N2 A0A182TPG6 A0A2A4IVE2 A0A2H1WUD0 A0A182V7K7 A0A067R0F9 A0A182XID0 A0A182HTB5 A0A182LFG8 Q7QBX7 A0A182Y799 A0A2H1WJP8 A0A232EX80 A0A232F2D1 B0W6G8 A0A067QLF8 A0A2H1VT20 A0A2J7PUL3 A0A139WJS1 A0A212FB51 A0A2A4K633 A0A182PA26 A0A1Y1KBU1 A0A1L8DPS2 A0A1L8DQ28 K7J8S8 A0A182MKD5 A0A0J9TKN6 A0A1L8DPY1 A0A1L8DPM5 A0A0C9RRQ5 A0A2A4JCC5 A0A3F2YX72 A0A034VW56 A0A0Q5WBH0 A0A194QUC1 A0A2W1B637 B4HWR7 A0A0J9R159 A0A1W7R7P1 Q16MD1 A0A182VQ94 A0A1A9UTK2 A0A0L7LE27 A0A0R1DRH5 B3N5D9 M9MRM3 A0A0L0C6A4 A0A194PXA6 B0W003 A0A226E1Q5 W8C916 B4P0Q9 A0A2H1X0E5 A0A2H1VWQ8 Q16F27 Q9VKT9 A0A2J7PY44 A0A1B0B8Y7 A0A194QNR0 A0A194PWK6 M9PCU5 A0A1W4W6G4 A0A2H1WZN4 H9J3W1 A0A0K8W418 A0A1S4FVZ7 A0A1D2MRG2 Q17BM4 V5GWI8 A0A067RHQ8 A0A1W4UT82 A0A2A4K6W6 A0A1Y1MRZ1 A0A1A9XQ71 A0A1B2M4Y7 A0A0L7LCZ5 A0A1W4UT02 A0A1B0CSN9
A0A2W1BS78 A0A067RCF7 A0A2J7QW32 A0A0T6BEX7 A0A194PMH9 A0A084W6W2 A0A194R2Y3 W5JF51 D2A0X6 A0A182JMM3 A0A194PL00 A0A182RCW3 A0A182KCP4 A0A212EKP9 A0A182FR70 A0A182H3N2 A0A182TPG6 A0A2A4IVE2 A0A2H1WUD0 A0A182V7K7 A0A067R0F9 A0A182XID0 A0A182HTB5 A0A182LFG8 Q7QBX7 A0A182Y799 A0A2H1WJP8 A0A232EX80 A0A232F2D1 B0W6G8 A0A067QLF8 A0A2H1VT20 A0A2J7PUL3 A0A139WJS1 A0A212FB51 A0A2A4K633 A0A182PA26 A0A1Y1KBU1 A0A1L8DPS2 A0A1L8DQ28 K7J8S8 A0A182MKD5 A0A0J9TKN6 A0A1L8DPY1 A0A1L8DPM5 A0A0C9RRQ5 A0A2A4JCC5 A0A3F2YX72 A0A034VW56 A0A0Q5WBH0 A0A194QUC1 A0A2W1B637 B4HWR7 A0A0J9R159 A0A1W7R7P1 Q16MD1 A0A182VQ94 A0A1A9UTK2 A0A0L7LE27 A0A0R1DRH5 B3N5D9 M9MRM3 A0A0L0C6A4 A0A194PXA6 B0W003 A0A226E1Q5 W8C916 B4P0Q9 A0A2H1X0E5 A0A2H1VWQ8 Q16F27 Q9VKT9 A0A2J7PY44 A0A1B0B8Y7 A0A194QNR0 A0A194PWK6 M9PCU5 A0A1W4W6G4 A0A2H1WZN4 H9J3W1 A0A0K8W418 A0A1S4FVZ7 A0A1D2MRG2 Q17BM4 V5GWI8 A0A067RHQ8 A0A1W4UT82 A0A2A4K6W6 A0A1Y1MRZ1 A0A1A9XQ71 A0A1B2M4Y7 A0A0L7LCZ5 A0A1W4UT02 A0A1B0CSN9
PDB
1K8Q
E-value=5.84987e-61,
Score=594
Ontologies
KEGG
GO
Topology
SignalP
Position: 1 - 16,
Likelihood: 0.861495
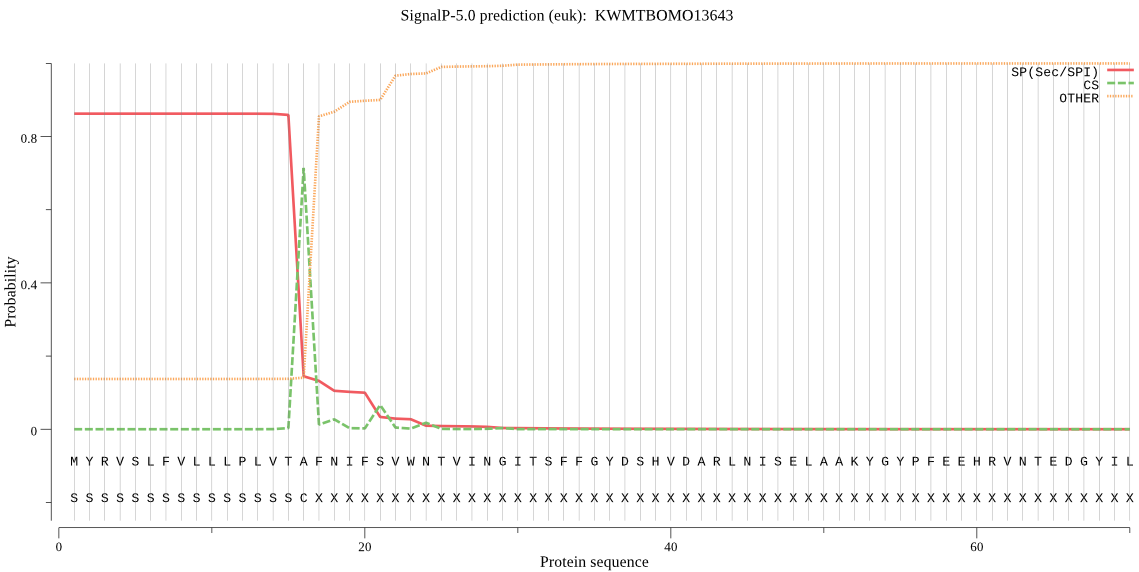
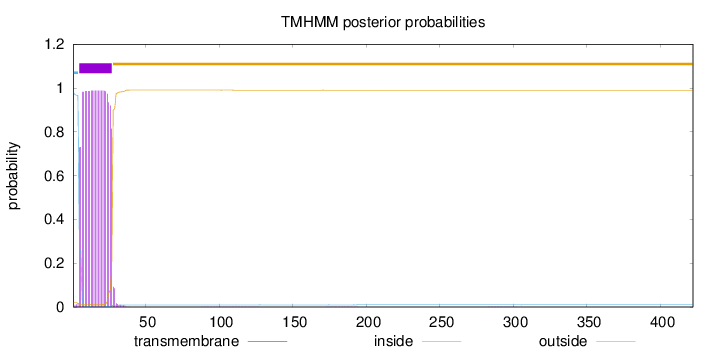
Length:
422
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.2044
Exp number, first 60 AAs:
22.16599
Total prob of N-in:
0.97933
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 422
Population Genetic Test Statistics
Pi
240.746035
Theta
188.288905
Tajima's D
0.810822
CLR
0.417313
CSRT
0.608419579021049
Interpretation
Uncertain
