Gene
KWMTBOMO13628 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011064
Annotation
PREDICTED:_delta-1-pyrroline-5-carboxylate_synthase_isoform_X1_[Papilio_polytes]
Full name
Delta-1-pyrroline-5-carboxylate synthase
Location in the cell
Cytoplasmic Reliability : 1.617 Mitochondrial Reliability : 2.148
Sequence
CDS
ATGATGTCGGATGTCGATGGTATTTACAACAAACCGCCCTGGGAAGACGGCGCGCGGATGATGCACACATTCACTTCCAACGAGATGGACCAGGTTCAGTTTGGGAAGAAATCTAAGGTGGGAACAGGTGGTATGGATTCCAAAGTCAATGCCGCCACTTGGGCCATGGCGCGTGGAGTCAGTGTGGTCATCTGTAACGGTATGCAAGAGAAGGCGATAAAAACCATTATTAGCGGTAGAAAAGTGGGAACATTTTTTACGGACTATGTTACTAGCTCCACCACTTCTGTCGATGAATTGGCTGAGAATGCTCGTACTGGTGGTCGAGCTCTTCAGCAACTGACTCCTCTAGACAGAGCGTCTGCTATTCACTCGTTAGCAGATTTACTATTGAGCAGACAAGATCAGATATTAGAAGCAAACAACAAAGATTTGGAGGAAGCTACCAAAAATGGTGTAGCGAAACCATTGCTAAACAGGCTGTCGCTCAGCCAAAGTAAATTAAAAACTTTATCGGTAGGATTGAAACAAATAGCTGATTCAAGTTACGAAAACGTCGGCAGGGTGCTTAGGAAGACCAAATTGGCTGAAAATCTCATTCTAAGACAAGTTACTGTACCTATAGGTGTATTGCTAGTTATATTTGAATCTAGGCCTGATTCGCTACCACAAGTCGCGGCCTTAGCTATGGCATCAGCTAACGGTCTTTTACTCAAAGGCGGAAAGGAAGCAGCGTACTCGAATAAAGCATTAATGGGGCTAGTTAAGGAATCTCTTAAAACGGTCGGAGCCGAAGATGCTATTTCTTTGATTTCGACACGGGAAGAAATAAGTGATTTGTTAGCTATGGACAAACACATTGATTTGATAATACCGAGAGGATCGTCGGAACTTGTTCAAAGTATACAGAAACAGTCGCAGCACATTCCCGTATTGGGACACGCTGAAGGAATATGTCATGTGTATTTAGATAAAGACGCCGACACAACTAAAGCTTTGAAGATAGTACGCGATGCGAAATGCGATTATCCGGCCGCTTGTAATGCTATGGAAACCTTACTTGTGCACGAGGACCACATCTCAGGATCATTGTTCACTGACGTTTGTAATATGTTGAAGAACGAAGGTGTGATAATCCACGCCGGTCCAAAACTAGCGAGACAGTTAACCTTCGGTCCTGCGCCGGCGAGGACCATGCGTCATGAGTACGGAGCCTTGGAGTGTTGTATTGAAGTCGTCAAGGATCTGGATGAAGCTGTTGATCATGTCCACAAATTCGGCAGCTCGCACACTGACGTTATTGTCACGGAAAACGAAAACGCAGCAAAACGGTTCTTGACGGCAGTGGATAGCGCCTGTGTTTTCCACAACGTGTCGTCTAGATTCGCTGATGGCTTCAGATTTGGTTTAGGGGCGGAAGTCGGAATATCGACTGCAAGGATACACGCAAGAGGTCCAGTCGGTGTAGAAGGTCTGTTAACAACGAAATGGATATTGGAGGGAACTGATCACACTGCAGCCGAGTTCAATGAGGGCAAACGAAACTGGATACACGAGAAAATGCCAGTCGCATGA
Protein
MMSDVDGIYNKPPWEDGARMMHTFTSNEMDQVQFGKKSKVGTGGMDSKVNAATWAMARGVSVVICNGMQEKAIKTIISGRKVGTFFTDYVTSSTTSVDELAENARTGGRALQQLTPLDRASAIHSLADLLLSRQDQILEANNKDLEEATKNGVAKPLLNRLSLSQSKLKTLSVGLKQIADSSYENVGRVLRKTKLAENLILRQVTVPIGVLLVIFESRPDSLPQVAALAMASANGLLLKGGKEAAYSNKALMGLVKESLKTVGAEDAISLISTREEISDLLAMDKHIDLIIPRGSSELVQSIQKQSQHIPVLGHAEGICHVYLDKDADTTKALKIVRDAKCDYPAACNAMETLLVHEDHISGSLFTDVCNMLKNEGVIIHAGPKLARQLTFGPAPARTMRHEYGALECCIEVVKDLDEAVDHVHKFGSSHTDVIVTENENAAKRFLTAVDSACVFHNVSSRFADGFRFGLGAEVGISTARIHARGPVGVEGLLTTKWILEGTDHTAAEFNEGKRNWIHEKMPVA
Summary
Catalytic Activity
ATP + L-glutamate = ADP + L-glutamyl 5-phosphate
L-glutamate 5-semialdehyde + NADP(+) + phosphate = H(+) + L-glutamyl 5-phosphate + NADPH
L-glutamate 5-semialdehyde + NADP(+) + phosphate = H(+) + L-glutamyl 5-phosphate + NADPH
Similarity
In the C-terminal section; belongs to the gamma-glutamyl phosphate reductase family.
In the N-terminal section; belongs to the glutamate 5-kinase family.
In the N-terminal section; belongs to the glutamate 5-kinase family.
Uniprot
H9JNG1
A0A3S2NZJ0
A0A0N0PB61
A0A194PRA6
A0A212EQ50
A0A2H1VTG9
+ More
A0A0L0CA23 A0A1I8NYW2 W8BEN1 T1PC19 Q29DN1 D3TQ43 B4PI54 A0A0J9S007 A0A1B0FQZ3 B4IAX9 A0A1L8EDS1 Q9VNW6 A0A1W4VBE6 A0A0P9AMM0 B3MAJ3 B3NJ19 A0A1Q3FKP5 A0A1Q3FKM4 A0A1Q3FKQ6 A0A0A1X3I7 B4L010 B4N4G2 A0A0K8VLQ1 A0A034WHU4 B0X119 Q174N2 D6WB12 A0A3B0J4E6 A0A182GWK4 A0A3B0JZ58 A0A2A4JN75 B4LHT5 A0A2A4JNZ9 A0A0M4EL01 U5ERF6 A0A0M4EJR2 B4J0Q4 A0A2M4BDG8 A0A2W1BSE8 A0A2M4BEU6 A0A2M4BER7 A0A2M4BER9 A0A0T6B002 A0A2M4A2W4 A0A2M4BET7 A0A182FRI6 A0A182TJE6 A0A2M4CFX4 A0A182P967 A0A182RGV7 A0A182V8L0 A0A182VWS8 A0A182SXN5 Q7QC97 A0A1J1HN75 W5J3P0 A0A182XHW0 A0A182IPP9 A0A2M4CFZ6 A0A182LEX3 A0A2M3YY67 A0A182HSU2 A0A182NRQ5 A0A182YRH0 A0A2M3YY87 A0A084VAR3 A0A182QGH8 A0A182JZU9 A0A0C5LC29 A0A1W4WY76 A0A1B0CT44 A0A336LQW4 A0A1L8E2V4 A0A232EWC8 K7INN6 A0A2A3EQN3 V9IDN8 V9ICB8 A0A1S6KZB3 A0A1B6CSU1 A0A0L7RBT4 A0A088A822 A0A023F5I9 A0A0C9QX27 A0A154PN15 J9JKH0 A0A2S2P4C9 A0A2H8TF50 N6TSS3 E0VJ34 A0A026WDH3 A0A224X845 A0A310SDF6 A0A1Y1LVX2
A0A0L0CA23 A0A1I8NYW2 W8BEN1 T1PC19 Q29DN1 D3TQ43 B4PI54 A0A0J9S007 A0A1B0FQZ3 B4IAX9 A0A1L8EDS1 Q9VNW6 A0A1W4VBE6 A0A0P9AMM0 B3MAJ3 B3NJ19 A0A1Q3FKP5 A0A1Q3FKM4 A0A1Q3FKQ6 A0A0A1X3I7 B4L010 B4N4G2 A0A0K8VLQ1 A0A034WHU4 B0X119 Q174N2 D6WB12 A0A3B0J4E6 A0A182GWK4 A0A3B0JZ58 A0A2A4JN75 B4LHT5 A0A2A4JNZ9 A0A0M4EL01 U5ERF6 A0A0M4EJR2 B4J0Q4 A0A2M4BDG8 A0A2W1BSE8 A0A2M4BEU6 A0A2M4BER7 A0A2M4BER9 A0A0T6B002 A0A2M4A2W4 A0A2M4BET7 A0A182FRI6 A0A182TJE6 A0A2M4CFX4 A0A182P967 A0A182RGV7 A0A182V8L0 A0A182VWS8 A0A182SXN5 Q7QC97 A0A1J1HN75 W5J3P0 A0A182XHW0 A0A182IPP9 A0A2M4CFZ6 A0A182LEX3 A0A2M3YY67 A0A182HSU2 A0A182NRQ5 A0A182YRH0 A0A2M3YY87 A0A084VAR3 A0A182QGH8 A0A182JZU9 A0A0C5LC29 A0A1W4WY76 A0A1B0CT44 A0A336LQW4 A0A1L8E2V4 A0A232EWC8 K7INN6 A0A2A3EQN3 V9IDN8 V9ICB8 A0A1S6KZB3 A0A1B6CSU1 A0A0L7RBT4 A0A088A822 A0A023F5I9 A0A0C9QX27 A0A154PN15 J9JKH0 A0A2S2P4C9 A0A2H8TF50 N6TSS3 E0VJ34 A0A026WDH3 A0A224X845 A0A310SDF6 A0A1Y1LVX2
Pubmed
19121390
26354079
22118469
26108605
24495485
25315136
+ More
15632085 17994087 20353571 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 25348373 17510324 18362917 19820115 26483478 28756777 12364791 14747013 17210077 20920257 23761445 20966253 25244985 24438588 25450565 28648823 20075255 24330026 25474469 23537049 20566863 24508170 30249741 28004739
15632085 17994087 20353571 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 25348373 17510324 18362917 19820115 26483478 28756777 12364791 14747013 17210077 20920257 23761445 20966253 25244985 24438588 25450565 28648823 20075255 24330026 25474469 23537049 20566863 24508170 30249741 28004739
EMBL
BABH01019270
BABH01019271
BABH01019272
RSAL01000015
RVE53123.1
KQ461153
+ More
KPJ08668.1 KQ459595 KPI95842.1 AGBW02013317 OWR43609.1 ODYU01004343 SOQ44117.1 JRES01000704 KNC29065.1 GAMC01014894 JAB91661.1 KA646316 AFP60945.1 CH379070 EAL30383.2 EZ423545 ADD19821.1 CM000159 EDW95502.1 CM002912 KMZ01138.1 CCAG010001599 CH480826 EDW44442.1 GFDG01001995 JAV16804.1 AE014296 AY069110 AAF51799.1 AAL39255.1 CH902618 KPU79024.1 EDV41280.1 CH954178 EDV52736.1 GFDL01006971 JAV28074.1 GFDL01006991 JAV28054.1 GFDL01006961 JAV28084.1 GBXI01008408 JAD05884.1 CH933809 EDW19045.1 CH964095 EDW79036.2 GDHF01012515 JAI39799.1 GAKP01005035 JAC53917.1 DS232250 EDS38411.1 CH477408 EAT41523.1 KQ971311 EEZ98981.2 OUUW01000002 SPP76397.1 JXUM01019982 KQ560560 KXJ81895.1 SPP76398.1 NWSH01000935 PCG73487.1 CH940647 EDW70660.1 PCG73486.1 CP012525 ALC44951.1 GANO01002826 JAB57045.1 ALC45240.1 CH916366 EDV97909.1 GGFJ01001958 MBW51099.1 KZ149986 PZC75680.1 GGFJ01002423 MBW51564.1 GGFJ01002409 MBW51550.1 GGFJ01002408 MBW51549.1 LJIG01016401 KRT80713.1 GGFK01001806 MBW35127.1 GGFJ01002424 MBW51565.1 GGFL01000064 MBW64242.1 AAAB01008859 EAA07467.4 CVRI01000004 CRK87689.1 ADMH02002105 ETN58982.1 GGFL01000065 MBW64243.1 GGFM01000471 MBW21222.1 APCN01000361 GGFM01000466 MBW21217.1 ATLV01004137 KE524190 KFB35057.1 AXCN02000008 KJ802127 AJP75147.1 AJWK01027055 AJWK01027056 UFQT01000054 SSX19073.1 GFDF01001182 JAV12902.1 NNAY01001889 OXU22631.1 KZ288203 PBC33351.1 JR039595 AEY58772.1 JR039596 AEY58773.1 KC223160 AQT27063.1 GEDC01026095 GEDC01020712 GEDC01009703 GEDC01003316 JAS11203.1 JAS16586.1 JAS27595.1 JAS33982.1 KQ414617 KOC68265.1 GBBI01002284 JAC16428.1 GBYB01005227 JAG74994.1 KQ434977 KZC12854.1 ABLF02026058 ABLF02026061 GGMR01011615 MBY24234.1 GFXV01000932 MBW12737.1 APGK01053233 KB741223 KB632273 ENN72325.1 ERL91075.1 AAZO01002758 DS235219 EEB13390.1 KK107274 QOIP01000008 EZA53716.1 RLU19584.1 GFTR01007911 JAW08515.1 KQ773785 OAD52349.1 GEZM01049917 JAV75756.1
KPJ08668.1 KQ459595 KPI95842.1 AGBW02013317 OWR43609.1 ODYU01004343 SOQ44117.1 JRES01000704 KNC29065.1 GAMC01014894 JAB91661.1 KA646316 AFP60945.1 CH379070 EAL30383.2 EZ423545 ADD19821.1 CM000159 EDW95502.1 CM002912 KMZ01138.1 CCAG010001599 CH480826 EDW44442.1 GFDG01001995 JAV16804.1 AE014296 AY069110 AAF51799.1 AAL39255.1 CH902618 KPU79024.1 EDV41280.1 CH954178 EDV52736.1 GFDL01006971 JAV28074.1 GFDL01006991 JAV28054.1 GFDL01006961 JAV28084.1 GBXI01008408 JAD05884.1 CH933809 EDW19045.1 CH964095 EDW79036.2 GDHF01012515 JAI39799.1 GAKP01005035 JAC53917.1 DS232250 EDS38411.1 CH477408 EAT41523.1 KQ971311 EEZ98981.2 OUUW01000002 SPP76397.1 JXUM01019982 KQ560560 KXJ81895.1 SPP76398.1 NWSH01000935 PCG73487.1 CH940647 EDW70660.1 PCG73486.1 CP012525 ALC44951.1 GANO01002826 JAB57045.1 ALC45240.1 CH916366 EDV97909.1 GGFJ01001958 MBW51099.1 KZ149986 PZC75680.1 GGFJ01002423 MBW51564.1 GGFJ01002409 MBW51550.1 GGFJ01002408 MBW51549.1 LJIG01016401 KRT80713.1 GGFK01001806 MBW35127.1 GGFJ01002424 MBW51565.1 GGFL01000064 MBW64242.1 AAAB01008859 EAA07467.4 CVRI01000004 CRK87689.1 ADMH02002105 ETN58982.1 GGFL01000065 MBW64243.1 GGFM01000471 MBW21222.1 APCN01000361 GGFM01000466 MBW21217.1 ATLV01004137 KE524190 KFB35057.1 AXCN02000008 KJ802127 AJP75147.1 AJWK01027055 AJWK01027056 UFQT01000054 SSX19073.1 GFDF01001182 JAV12902.1 NNAY01001889 OXU22631.1 KZ288203 PBC33351.1 JR039595 AEY58772.1 JR039596 AEY58773.1 KC223160 AQT27063.1 GEDC01026095 GEDC01020712 GEDC01009703 GEDC01003316 JAS11203.1 JAS16586.1 JAS27595.1 JAS33982.1 KQ414617 KOC68265.1 GBBI01002284 JAC16428.1 GBYB01005227 JAG74994.1 KQ434977 KZC12854.1 ABLF02026058 ABLF02026061 GGMR01011615 MBY24234.1 GFXV01000932 MBW12737.1 APGK01053233 KB741223 KB632273 ENN72325.1 ERL91075.1 AAZO01002758 DS235219 EEB13390.1 KK107274 QOIP01000008 EZA53716.1 RLU19584.1 GFTR01007911 JAW08515.1 KQ773785 OAD52349.1 GEZM01049917 JAV75756.1
Proteomes
UP000005204
UP000283053
UP000053240
UP000053268
UP000007151
UP000037069
+ More
UP000095300 UP000095301 UP000001819 UP000002282 UP000092444 UP000001292 UP000000803 UP000192221 UP000007801 UP000008711 UP000009192 UP000007798 UP000002320 UP000008820 UP000007266 UP000268350 UP000069940 UP000249989 UP000218220 UP000008792 UP000092553 UP000001070 UP000069272 UP000075902 UP000075885 UP000075900 UP000075903 UP000075920 UP000075901 UP000007062 UP000183832 UP000000673 UP000076407 UP000075880 UP000075882 UP000075840 UP000075884 UP000076408 UP000030765 UP000075886 UP000075881 UP000192223 UP000092461 UP000215335 UP000002358 UP000242457 UP000053825 UP000005203 UP000076502 UP000007819 UP000019118 UP000030742 UP000009046 UP000053097 UP000279307
UP000095300 UP000095301 UP000001819 UP000002282 UP000092444 UP000001292 UP000000803 UP000192221 UP000007801 UP000008711 UP000009192 UP000007798 UP000002320 UP000008820 UP000007266 UP000268350 UP000069940 UP000249989 UP000218220 UP000008792 UP000092553 UP000001070 UP000069272 UP000075902 UP000075885 UP000075900 UP000075903 UP000075920 UP000075901 UP000007062 UP000183832 UP000000673 UP000076407 UP000075880 UP000075882 UP000075840 UP000075884 UP000076408 UP000030765 UP000075886 UP000075881 UP000192223 UP000092461 UP000215335 UP000002358 UP000242457 UP000053825 UP000005203 UP000076502 UP000007819 UP000019118 UP000030742 UP000009046 UP000053097 UP000279307
PRIDE
Interpro
IPR000965
GPR_dom
+ More
IPR016162 Ald_DH_N
IPR015590 Aldehyde_DH_dom
IPR016161 Ald_DH/histidinol_DH
IPR005715 Glu_5kinase/COase_Synthase
IPR001048 Asp/Glu/Uridylate_kinase
IPR005766 P5_carboxy_syn
IPR036393 AceGlu_kinase-like_sf
IPR041744 G5K_ProBA
IPR016163 Ald_DH_C
IPR001057 Glu/AcGlu_kinase
IPR019797 Glutamate_5-kinase_CS
IPR020593 G-glutamylP_reductase_CS
IPR016162 Ald_DH_N
IPR015590 Aldehyde_DH_dom
IPR016161 Ald_DH/histidinol_DH
IPR005715 Glu_5kinase/COase_Synthase
IPR001048 Asp/Glu/Uridylate_kinase
IPR005766 P5_carboxy_syn
IPR036393 AceGlu_kinase-like_sf
IPR041744 G5K_ProBA
IPR016163 Ald_DH_C
IPR001057 Glu/AcGlu_kinase
IPR019797 Glutamate_5-kinase_CS
IPR020593 G-glutamylP_reductase_CS
Gene 3D
ProteinModelPortal
H9JNG1
A0A3S2NZJ0
A0A0N0PB61
A0A194PRA6
A0A212EQ50
A0A2H1VTG9
+ More
A0A0L0CA23 A0A1I8NYW2 W8BEN1 T1PC19 Q29DN1 D3TQ43 B4PI54 A0A0J9S007 A0A1B0FQZ3 B4IAX9 A0A1L8EDS1 Q9VNW6 A0A1W4VBE6 A0A0P9AMM0 B3MAJ3 B3NJ19 A0A1Q3FKP5 A0A1Q3FKM4 A0A1Q3FKQ6 A0A0A1X3I7 B4L010 B4N4G2 A0A0K8VLQ1 A0A034WHU4 B0X119 Q174N2 D6WB12 A0A3B0J4E6 A0A182GWK4 A0A3B0JZ58 A0A2A4JN75 B4LHT5 A0A2A4JNZ9 A0A0M4EL01 U5ERF6 A0A0M4EJR2 B4J0Q4 A0A2M4BDG8 A0A2W1BSE8 A0A2M4BEU6 A0A2M4BER7 A0A2M4BER9 A0A0T6B002 A0A2M4A2W4 A0A2M4BET7 A0A182FRI6 A0A182TJE6 A0A2M4CFX4 A0A182P967 A0A182RGV7 A0A182V8L0 A0A182VWS8 A0A182SXN5 Q7QC97 A0A1J1HN75 W5J3P0 A0A182XHW0 A0A182IPP9 A0A2M4CFZ6 A0A182LEX3 A0A2M3YY67 A0A182HSU2 A0A182NRQ5 A0A182YRH0 A0A2M3YY87 A0A084VAR3 A0A182QGH8 A0A182JZU9 A0A0C5LC29 A0A1W4WY76 A0A1B0CT44 A0A336LQW4 A0A1L8E2V4 A0A232EWC8 K7INN6 A0A2A3EQN3 V9IDN8 V9ICB8 A0A1S6KZB3 A0A1B6CSU1 A0A0L7RBT4 A0A088A822 A0A023F5I9 A0A0C9QX27 A0A154PN15 J9JKH0 A0A2S2P4C9 A0A2H8TF50 N6TSS3 E0VJ34 A0A026WDH3 A0A224X845 A0A310SDF6 A0A1Y1LVX2
A0A0L0CA23 A0A1I8NYW2 W8BEN1 T1PC19 Q29DN1 D3TQ43 B4PI54 A0A0J9S007 A0A1B0FQZ3 B4IAX9 A0A1L8EDS1 Q9VNW6 A0A1W4VBE6 A0A0P9AMM0 B3MAJ3 B3NJ19 A0A1Q3FKP5 A0A1Q3FKM4 A0A1Q3FKQ6 A0A0A1X3I7 B4L010 B4N4G2 A0A0K8VLQ1 A0A034WHU4 B0X119 Q174N2 D6WB12 A0A3B0J4E6 A0A182GWK4 A0A3B0JZ58 A0A2A4JN75 B4LHT5 A0A2A4JNZ9 A0A0M4EL01 U5ERF6 A0A0M4EJR2 B4J0Q4 A0A2M4BDG8 A0A2W1BSE8 A0A2M4BEU6 A0A2M4BER7 A0A2M4BER9 A0A0T6B002 A0A2M4A2W4 A0A2M4BET7 A0A182FRI6 A0A182TJE6 A0A2M4CFX4 A0A182P967 A0A182RGV7 A0A182V8L0 A0A182VWS8 A0A182SXN5 Q7QC97 A0A1J1HN75 W5J3P0 A0A182XHW0 A0A182IPP9 A0A2M4CFZ6 A0A182LEX3 A0A2M3YY67 A0A182HSU2 A0A182NRQ5 A0A182YRH0 A0A2M3YY87 A0A084VAR3 A0A182QGH8 A0A182JZU9 A0A0C5LC29 A0A1W4WY76 A0A1B0CT44 A0A336LQW4 A0A1L8E2V4 A0A232EWC8 K7INN6 A0A2A3EQN3 V9IDN8 V9ICB8 A0A1S6KZB3 A0A1B6CSU1 A0A0L7RBT4 A0A088A822 A0A023F5I9 A0A0C9QX27 A0A154PN15 J9JKH0 A0A2S2P4C9 A0A2H8TF50 N6TSS3 E0VJ34 A0A026WDH3 A0A224X845 A0A310SDF6 A0A1Y1LVX2
PDB
2H5G
E-value=1.23617e-147,
Score=1342
Ontologies
PATHWAY
GO
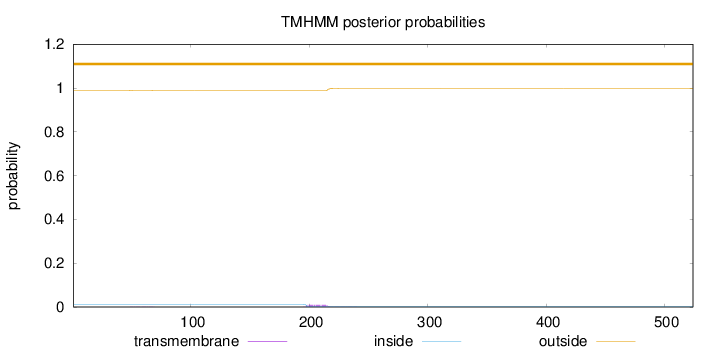
Topology
Length:
524
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.19709
Exp number, first 60 AAs:
0.00542
Total prob of N-in:
0.01148
outside
1 - 524
Population Genetic Test Statistics
Pi
28.909404
Theta
19.184357
Tajima's D
0.305249
CLR
1.900067
CSRT
0.469226538673066
Interpretation
Uncertain
