Pre Gene Modal
BGIBMGA011062
Annotation
LSM1-like_protein_[Bombyx_mori]
Full name
U6 snRNA-associated Sm-like protein LSm1
Location in the cell
Cytoplasmic Reliability : 1.427 Nuclear Reliability : 1.244
Sequence
CDS
ATGTCTAGTAATACAAACCCTTTAGCTGGGACGGCGCATCTTCTCGACGAATTAGACAAGAAACTAATGGTGTTGCTGCGTGACGGAAGGACGCTTATCGGGTATTTACGCTGCGTGGATCAATTCGCTAATCTAGTGCTCCACAAGACAATAGAACGAATACACGTTGGCAAAGAATACGGTGATATTCCGAGAGGCATTTTCATTGTTAGAGGAGAGAATGTTGTTCTACTTGGTGAAATAGACAAAGACAAAGAAGAAAATCTCCCTCTAACGGAAGTATCAGTAGATGAAATACTAGATGCTCAAAGAAGAGAACAAGATGCGAAGATTGATCAACAAAAATTGTTATCAAAAGCTTTAAAAGAAAGAGGGCTTAATTTACTAGCAGATCTTGGACACGATGATATGTTTTAA
Protein
MSSNTNPLAGTAHLLDELDKKLMVLLRDGRTLIGYLRCVDQFANLVLHKTIERIHVGKEYGDIPRGIFIVRGENVVLLGEIDKDKEENLPLTEVSVDEILDAQRREQDAKIDQQKLLSKALKERGLNLLADLGHDDMF
Summary
Description
Probably involved with other LSm subunits in the general process of degradation of mRNAs.
Subunit
LSm subunits form a heteromer with a donut shape.
Similarity
Belongs to the snRNP Sm proteins family.
Uniprot
Q1HQ51
A0A3S2MAY4
A0A2A4JPX3
A0A2W1BV98
A0A0N1IBU3
I4DKE1
+ More
A0A2H1VYC2 A0A0L7L8H1 S4PDA2 A0A212EQ38 H9JNF9 A0A1E1WST2 A0A1Y1N6W9 A0A0L7RK61 K7J9P3 V9IL93 A0A088AQN3 A0A0C9R6G9 A0A067R7S7 A0A2J7QCJ0 N6TSW2 A0A2A3EGX0 A0A182GUR6 A0A1B6GJ98 E2B862 A0A023EG51 A0A1Q3G367 Q17P42 A0A1L8DGM8 A0A1B6MSM3 B0WRR5 A0A1B0DGC5 A0A1B6L6G9 A0A0K8TPD1 D7EI70 A0A2M3ZD33 A0A2M4AYL9 A0A2M4C360 W5J1H3 A0A182FQP5 A0A069DPD0 A0A0V0G929 E0VYP1 T1GW79 A0A084VQL0 A0A182J522 A0A0P4VP74 R4FL32 Q4V6R8 A0A023FAS4 A0A336LP47 A0A2R7WMS2 U5EWI4 A0A0T6AZF8 A0A182QB70 E2ARW6 A0A0A9XH16 A0A1B6DJZ1 A0A182LX64 A0A182R8V5 A0A182Y3Q0 A0A195EJI4 A0A151ILB4 F4X418 A0A195BGV0 A0A195FM71 A0A158NJL6 A0A151X6I3 A0A182NRX0 E9JC43 A0A182PSL9 A0A182VZJ1 A0A182US35 A0A182JZH7 A0A182TE41 A0A182LA02 A0A182WSQ2 Q7PY05 A0A182HLK4 T1DNB8 A0A154NZF6 A0A0L0BUI5 B4MNX6 A0A0K8VZK0 A0A034V1C6 A0A1I8NGT4 A0A0A1WI70 B4LMQ7 B4KNC1 A0A0M3QUM0 B3MDT1 W8BVL5 A0A1I8PGW4 B3NJP1 A0A3B0JTK9 Q28X08 B4H4V3 A0A1W4UMK3 B4P9G6 B4I7L2
A0A2H1VYC2 A0A0L7L8H1 S4PDA2 A0A212EQ38 H9JNF9 A0A1E1WST2 A0A1Y1N6W9 A0A0L7RK61 K7J9P3 V9IL93 A0A088AQN3 A0A0C9R6G9 A0A067R7S7 A0A2J7QCJ0 N6TSW2 A0A2A3EGX0 A0A182GUR6 A0A1B6GJ98 E2B862 A0A023EG51 A0A1Q3G367 Q17P42 A0A1L8DGM8 A0A1B6MSM3 B0WRR5 A0A1B0DGC5 A0A1B6L6G9 A0A0K8TPD1 D7EI70 A0A2M3ZD33 A0A2M4AYL9 A0A2M4C360 W5J1H3 A0A182FQP5 A0A069DPD0 A0A0V0G929 E0VYP1 T1GW79 A0A084VQL0 A0A182J522 A0A0P4VP74 R4FL32 Q4V6R8 A0A023FAS4 A0A336LP47 A0A2R7WMS2 U5EWI4 A0A0T6AZF8 A0A182QB70 E2ARW6 A0A0A9XH16 A0A1B6DJZ1 A0A182LX64 A0A182R8V5 A0A182Y3Q0 A0A195EJI4 A0A151ILB4 F4X418 A0A195BGV0 A0A195FM71 A0A158NJL6 A0A151X6I3 A0A182NRX0 E9JC43 A0A182PSL9 A0A182VZJ1 A0A182US35 A0A182JZH7 A0A182TE41 A0A182LA02 A0A182WSQ2 Q7PY05 A0A182HLK4 T1DNB8 A0A154NZF6 A0A0L0BUI5 B4MNX6 A0A0K8VZK0 A0A034V1C6 A0A1I8NGT4 A0A0A1WI70 B4LMQ7 B4KNC1 A0A0M3QUM0 B3MDT1 W8BVL5 A0A1I8PGW4 B3NJP1 A0A3B0JTK9 Q28X08 B4H4V3 A0A1W4UMK3 B4P9G6 B4I7L2
Pubmed
28756777
26354079
22651552
26227816
23622113
22118469
+ More
19121390 28004739 20075255 24845553 23537049 26483478 20798317 24945155 17510324 26369729 18362917 19820115 20920257 23761445 26334808 20566863 24438588 27129103 25474469 25401762 26823975 25244985 21719571 21347285 21282665 20966253 12364791 14747013 17210077 26108605 17994087 25348373 25315136 25830018 24495485 15632085 17550304
19121390 28004739 20075255 24845553 23537049 26483478 20798317 24945155 17510324 26369729 18362917 19820115 20920257 23761445 26334808 20566863 24438588 27129103 25474469 25401762 26823975 25244985 21719571 21347285 21282665 20966253 12364791 14747013 17210077 26108605 17994087 25348373 25315136 25830018 24495485 15632085 17550304
EMBL
DQ443201
ABF51290.1
RSAL01000002
RVE55024.1
RVE55077.1
NWSH01000935
+ More
PCG73483.1 KZ149986 PZC75683.1 KQ461153 KPJ08665.1 AK401759 KQ459595 BAM18381.1 KPI95844.1 ODYU01004884 SOQ45214.1 JTDY01002247 KOB71802.1 GAIX01003746 JAA88814.1 AGBW02013317 OWR43612.1 BABH01019270 GDQN01000994 JAT90060.1 GEZM01011086 JAV93654.1 KQ414578 KOC71198.1 AAZX01008083 JR052962 AEY61830.1 GBYB01008402 JAG78169.1 KK852643 KDR19580.1 NEVH01016289 PNF26297.1 APGK01053342 KB741223 KB630494 KB630981 ENN72365.1 ERL83629.1 ERL83978.1 KZ288253 PBC31015.1 JXUM01018890 KQ560525 KXJ81981.1 GECZ01027119 GECZ01007253 JAS42650.1 JAS62516.1 GL446284 EFN88119.1 GAPW01005798 JAC07800.1 GFDL01000793 JAV34252.1 CH477194 EAT48458.1 GFDF01008590 JAV05494.1 GEBQ01001084 JAT38893.1 DS232058 EDS33469.1 AJVK01059507 GEBQ01020659 JAT19318.1 GDAI01001602 JAI16001.1 DS497673 EFA13273.1 GGFM01005670 MBW26421.1 GGFK01012568 MBW45889.1 GGFJ01010593 MBW59734.1 ADMH02002181 ETN58022.1 GBGD01003412 JAC85477.1 GECL01001731 JAP04393.1 AAZO01006277 DS235845 EEB18497.1 CAQQ02153970 ATLV01015250 KE525004 KFB40254.1 GDKW01001801 JAI54794.1 ACPB03023383 GAHY01001593 JAA75917.1 BT022238 AAY54654.1 GBBI01000694 JAC18018.1 UFQT01000095 SSX19690.1 KK855095 PTY20799.1 GANO01001399 JAB58472.1 LJIG01022452 KRT80521.1 AXCN02000252 GL442192 EFN63815.1 GBHO01024332 GBHO01024330 GBRD01000620 GDHC01000076 JAG19272.1 JAG19274.1 JAG65201.1 JAQ18553.1 GEDC01011311 JAS25987.1 AXCM01000138 KQ978782 KYN28413.1 KQ977126 KYN05518.1 GL888624 EGI58951.1 KQ976490 KYM83446.1 KQ981424 KYN41795.1 ADTU01018046 KQ982482 KYQ55967.1 GL771706 EFZ09609.1 AAAB01008987 EAA00900.3 APCN01000909 GAMD01003025 JAA98565.1 KQ434777 KZC04260.1 JRES01001317 KNC23653.1 CH963848 EDW73815.1 GDHF01007976 JAI44338.1 GAKP01023373 JAC35585.1 GBXI01015956 GBXI01000253 JAC98335.1 JAD14039.1 CH940648 EDW60998.1 CH933808 EDW09974.1 CP012524 ALC40863.1 CH902619 EDV36466.1 GAMC01001200 JAC05356.1 CH954179 EDV55420.1 OUUW01000001 SPP74408.1 CM000071 EAL26508.1 CH479210 EDW32789.1 CM000158 EDW91286.1 CH480824 EDW56587.1
PCG73483.1 KZ149986 PZC75683.1 KQ461153 KPJ08665.1 AK401759 KQ459595 BAM18381.1 KPI95844.1 ODYU01004884 SOQ45214.1 JTDY01002247 KOB71802.1 GAIX01003746 JAA88814.1 AGBW02013317 OWR43612.1 BABH01019270 GDQN01000994 JAT90060.1 GEZM01011086 JAV93654.1 KQ414578 KOC71198.1 AAZX01008083 JR052962 AEY61830.1 GBYB01008402 JAG78169.1 KK852643 KDR19580.1 NEVH01016289 PNF26297.1 APGK01053342 KB741223 KB630494 KB630981 ENN72365.1 ERL83629.1 ERL83978.1 KZ288253 PBC31015.1 JXUM01018890 KQ560525 KXJ81981.1 GECZ01027119 GECZ01007253 JAS42650.1 JAS62516.1 GL446284 EFN88119.1 GAPW01005798 JAC07800.1 GFDL01000793 JAV34252.1 CH477194 EAT48458.1 GFDF01008590 JAV05494.1 GEBQ01001084 JAT38893.1 DS232058 EDS33469.1 AJVK01059507 GEBQ01020659 JAT19318.1 GDAI01001602 JAI16001.1 DS497673 EFA13273.1 GGFM01005670 MBW26421.1 GGFK01012568 MBW45889.1 GGFJ01010593 MBW59734.1 ADMH02002181 ETN58022.1 GBGD01003412 JAC85477.1 GECL01001731 JAP04393.1 AAZO01006277 DS235845 EEB18497.1 CAQQ02153970 ATLV01015250 KE525004 KFB40254.1 GDKW01001801 JAI54794.1 ACPB03023383 GAHY01001593 JAA75917.1 BT022238 AAY54654.1 GBBI01000694 JAC18018.1 UFQT01000095 SSX19690.1 KK855095 PTY20799.1 GANO01001399 JAB58472.1 LJIG01022452 KRT80521.1 AXCN02000252 GL442192 EFN63815.1 GBHO01024332 GBHO01024330 GBRD01000620 GDHC01000076 JAG19272.1 JAG19274.1 JAG65201.1 JAQ18553.1 GEDC01011311 JAS25987.1 AXCM01000138 KQ978782 KYN28413.1 KQ977126 KYN05518.1 GL888624 EGI58951.1 KQ976490 KYM83446.1 KQ981424 KYN41795.1 ADTU01018046 KQ982482 KYQ55967.1 GL771706 EFZ09609.1 AAAB01008987 EAA00900.3 APCN01000909 GAMD01003025 JAA98565.1 KQ434777 KZC04260.1 JRES01001317 KNC23653.1 CH963848 EDW73815.1 GDHF01007976 JAI44338.1 GAKP01023373 JAC35585.1 GBXI01015956 GBXI01000253 JAC98335.1 JAD14039.1 CH940648 EDW60998.1 CH933808 EDW09974.1 CP012524 ALC40863.1 CH902619 EDV36466.1 GAMC01001200 JAC05356.1 CH954179 EDV55420.1 OUUW01000001 SPP74408.1 CM000071 EAL26508.1 CH479210 EDW32789.1 CM000158 EDW91286.1 CH480824 EDW56587.1
Proteomes
UP000283053
UP000218220
UP000053240
UP000053268
UP000037510
UP000007151
+ More
UP000005204 UP000053825 UP000002358 UP000005203 UP000027135 UP000235965 UP000019118 UP000030742 UP000242457 UP000069940 UP000249989 UP000008237 UP000008820 UP000002320 UP000092462 UP000007266 UP000000673 UP000069272 UP000009046 UP000015102 UP000030765 UP000075880 UP000015103 UP000075886 UP000000311 UP000075883 UP000075900 UP000076408 UP000078492 UP000078542 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000075884 UP000075885 UP000075920 UP000075903 UP000075881 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000076502 UP000037069 UP000007798 UP000095301 UP000008792 UP000009192 UP000092553 UP000007801 UP000095300 UP000008711 UP000268350 UP000001819 UP000008744 UP000192221 UP000002282 UP000001292
UP000005204 UP000053825 UP000002358 UP000005203 UP000027135 UP000235965 UP000019118 UP000030742 UP000242457 UP000069940 UP000249989 UP000008237 UP000008820 UP000002320 UP000092462 UP000007266 UP000000673 UP000069272 UP000009046 UP000015102 UP000030765 UP000075880 UP000015103 UP000075886 UP000000311 UP000075883 UP000075900 UP000076408 UP000078492 UP000078542 UP000007755 UP000078540 UP000078541 UP000005205 UP000075809 UP000075884 UP000075885 UP000075920 UP000075903 UP000075881 UP000075902 UP000075882 UP000076407 UP000007062 UP000075840 UP000076502 UP000037069 UP000007798 UP000095301 UP000008792 UP000009192 UP000092553 UP000007801 UP000095300 UP000008711 UP000268350 UP000001819 UP000008744 UP000192221 UP000002282 UP000001292
Pfam
PF01423 LSM
SUPFAM
SSF50182
SSF50182
CDD
ProteinModelPortal
Q1HQ51
A0A3S2MAY4
A0A2A4JPX3
A0A2W1BV98
A0A0N1IBU3
I4DKE1
+ More
A0A2H1VYC2 A0A0L7L8H1 S4PDA2 A0A212EQ38 H9JNF9 A0A1E1WST2 A0A1Y1N6W9 A0A0L7RK61 K7J9P3 V9IL93 A0A088AQN3 A0A0C9R6G9 A0A067R7S7 A0A2J7QCJ0 N6TSW2 A0A2A3EGX0 A0A182GUR6 A0A1B6GJ98 E2B862 A0A023EG51 A0A1Q3G367 Q17P42 A0A1L8DGM8 A0A1B6MSM3 B0WRR5 A0A1B0DGC5 A0A1B6L6G9 A0A0K8TPD1 D7EI70 A0A2M3ZD33 A0A2M4AYL9 A0A2M4C360 W5J1H3 A0A182FQP5 A0A069DPD0 A0A0V0G929 E0VYP1 T1GW79 A0A084VQL0 A0A182J522 A0A0P4VP74 R4FL32 Q4V6R8 A0A023FAS4 A0A336LP47 A0A2R7WMS2 U5EWI4 A0A0T6AZF8 A0A182QB70 E2ARW6 A0A0A9XH16 A0A1B6DJZ1 A0A182LX64 A0A182R8V5 A0A182Y3Q0 A0A195EJI4 A0A151ILB4 F4X418 A0A195BGV0 A0A195FM71 A0A158NJL6 A0A151X6I3 A0A182NRX0 E9JC43 A0A182PSL9 A0A182VZJ1 A0A182US35 A0A182JZH7 A0A182TE41 A0A182LA02 A0A182WSQ2 Q7PY05 A0A182HLK4 T1DNB8 A0A154NZF6 A0A0L0BUI5 B4MNX6 A0A0K8VZK0 A0A034V1C6 A0A1I8NGT4 A0A0A1WI70 B4LMQ7 B4KNC1 A0A0M3QUM0 B3MDT1 W8BVL5 A0A1I8PGW4 B3NJP1 A0A3B0JTK9 Q28X08 B4H4V3 A0A1W4UMK3 B4P9G6 B4I7L2
A0A2H1VYC2 A0A0L7L8H1 S4PDA2 A0A212EQ38 H9JNF9 A0A1E1WST2 A0A1Y1N6W9 A0A0L7RK61 K7J9P3 V9IL93 A0A088AQN3 A0A0C9R6G9 A0A067R7S7 A0A2J7QCJ0 N6TSW2 A0A2A3EGX0 A0A182GUR6 A0A1B6GJ98 E2B862 A0A023EG51 A0A1Q3G367 Q17P42 A0A1L8DGM8 A0A1B6MSM3 B0WRR5 A0A1B0DGC5 A0A1B6L6G9 A0A0K8TPD1 D7EI70 A0A2M3ZD33 A0A2M4AYL9 A0A2M4C360 W5J1H3 A0A182FQP5 A0A069DPD0 A0A0V0G929 E0VYP1 T1GW79 A0A084VQL0 A0A182J522 A0A0P4VP74 R4FL32 Q4V6R8 A0A023FAS4 A0A336LP47 A0A2R7WMS2 U5EWI4 A0A0T6AZF8 A0A182QB70 E2ARW6 A0A0A9XH16 A0A1B6DJZ1 A0A182LX64 A0A182R8V5 A0A182Y3Q0 A0A195EJI4 A0A151ILB4 F4X418 A0A195BGV0 A0A195FM71 A0A158NJL6 A0A151X6I3 A0A182NRX0 E9JC43 A0A182PSL9 A0A182VZJ1 A0A182US35 A0A182JZH7 A0A182TE41 A0A182LA02 A0A182WSQ2 Q7PY05 A0A182HLK4 T1DNB8 A0A154NZF6 A0A0L0BUI5 B4MNX6 A0A0K8VZK0 A0A034V1C6 A0A1I8NGT4 A0A0A1WI70 B4LMQ7 B4KNC1 A0A0M3QUM0 B3MDT1 W8BVL5 A0A1I8PGW4 B3NJP1 A0A3B0JTK9 Q28X08 B4H4V3 A0A1W4UMK3 B4P9G6 B4I7L2
PDB
4C92
E-value=9.41682e-18,
Score=214
Ontologies
GO
Topology
Subcellular location
Cytoplasm
P-body
P-body
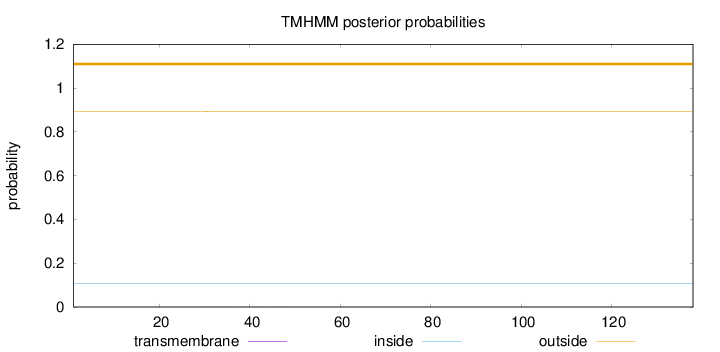
Length:
138
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0065
Exp number, first 60 AAs:
0.0065
Total prob of N-in:
0.10615
outside
1 - 138
Population Genetic Test Statistics
Pi
176.723463
Theta
162.486924
Tajima's D
0.446628
CLR
0.391834
CSRT
0.501824908754562
Interpretation
Uncertain
