Gene
KWMTBOMO13607
Pre Gene Modal
BGIBMGA006778
Annotation
Uncharacterized_protein_OBRU01_16492_[Operophtera_brumata]
Location in the cell
Nuclear Reliability : 2.881
Sequence
CDS
ATGGTAGAGAGCCAGGAGGTGCTTTCCCATTTAATAGGAAAGCGTCGTTACGTCAAAGGCTCACTTACAAGACTCTATAACCAGTTGGGAGAAACCGAAAGACTGTCTACGGAAACTATACAATGCCGCTTAGAGCGTTGTGTTGTAGCGTTCCGCGAATACGGCGATTATTGCGGCCGAATTCTGGCAACTAACCCGGCTGATGAAGAGGATTCAGGTGAGTACGATGACAAATATTTTGGCATCATGGCTTGCTACCAAGCTGAGCTTAACAAGCGAAAGGCCAATGCTCCGCCTAATCCGACACCCCAAAAAAGGCTTATAAAGTTACCTACCATAGAAATGCCGCAGTTCACGGGTCAATATTCGAAATATATCACATTTAGAGAAATGTTCAAATCAGTTATCGAAAATGATGTTTCTTTAGATGACATTCAAAAACTGCATTATTTGCGATCATTTTTAAAGGAAGAACCTTTAGATTTAATAATTAATTTGCCTCTTATAGGGTCCAGCTTCAAGGAAGCTCTGAGGTTACTGGACCAGAGATATTATAATAAATATAATATAATGACAGAGCTTTTGTCTACATTGCTAGATCTAGATGTTATTAGTAAACCTTCTCCTAGCAATATAAGACATTTTGTGTCAACAATTAAACAAACGTTGGCTGCCCTCAAAAATATGGAGGCGAAGGTAGATGAATGGGATCCAATTTTAATGTTGATTTTGACCCGCAAGTTAGATGAACGTACAGCACGCGAATACCAACTGGATCGGGACAACACAATGGACCCCACTGTGGACCAGCTTCTAGCGTTCCTTGAAAAAAGAGCGCTCGCTATGGAGAATGCTGGACAATCCGCAGCCAAGTTGTTTACCACCAAAGTAGTCAACGCAGTGACCGCTAAGGAGAAGCGAGGATGTTCAATTTATGTAGTGGACTGTATTATTAGGATGGGCAAGGAACTTGTTAAATAA
Protein
MVESQEVLSHLIGKRRYVKGSLTRLYNQLGETERLSTETIQCRLERCVVAFREYGDYCGRILATNPADEEDSGEYDDKYFGIMACYQAELNKRKANAPPNPTPQKRLIKLPTIEMPQFTGQYSKYITFREMFKSVIENDVSLDDIQKLHYLRSFLKEEPLDLIINLPLIGSSFKEALRLLDQRYYNKYNIMTELLSTLLDLDVISKPSPSNIRHFVSTIKQTLAALKNMEAKVDEWDPILMLILTRKLDERTAREYQLDRDNTMDPTVDQLLAFLEKRALAMENAGQSAAKLFTTKVVNAVTAKEKRGCSIYVVDCIIRMGKELVK
Summary
Uniprot
A0A0L7L2Y3
H9JTH4
A0A0L7LCW9
A0A1Y1JVV0
A0A1E1WTE8
A0A0L7LE91
+ More
A0A3S2P870 A0A1E1VXH0 A0A151ISI5 V5GPJ6 V5I888 A0A151IJK3 A0A1Y1N4T1 A0A151X752 A0A151IMM1 A0A0L7K424 A0A151IDJ5 A0A151XJI2 A0A151IDD4 A0A0L7LCW4 A0A151ITI6 A0A0J7K0K5 A0A2H1WH28 A0A151IAU9 A0A139W8E1 A0A0L7L2J2 A0A0L7KS35 A0A2H1WI74 A0A151IN79 A0A151ICE7 A0A0L7LB62 A0A2S2PFV9 A0A151JNR8 X1XTY9 A0A151JNU0 A0A0J7KLX1 A0A151JAJ1 A0A0N1IHU1 A0A151IRH4 A0A0L7KRI4 H9J9C1 A0A1Y1LJL9 H9JIA8 A0A195C7P8 A0A194RAG7 A0A151XF58 A0A2A4JL65 X1WNA5 A0A224XG39 A0A2S2NTG1 X1WSV3 S4NIJ2 A0A154P493 A0A1Y1KV73 J9LIE4 A0A151XET3 A0A023F0J1 A0A151JF33 J9KJ92 A0A2P8XCR5
A0A3S2P870 A0A1E1VXH0 A0A151ISI5 V5GPJ6 V5I888 A0A151IJK3 A0A1Y1N4T1 A0A151X752 A0A151IMM1 A0A0L7K424 A0A151IDJ5 A0A151XJI2 A0A151IDD4 A0A0L7LCW4 A0A151ITI6 A0A0J7K0K5 A0A2H1WH28 A0A151IAU9 A0A139W8E1 A0A0L7L2J2 A0A0L7KS35 A0A2H1WI74 A0A151IN79 A0A151ICE7 A0A0L7LB62 A0A2S2PFV9 A0A151JNR8 X1XTY9 A0A151JNU0 A0A0J7KLX1 A0A151JAJ1 A0A0N1IHU1 A0A151IRH4 A0A0L7KRI4 H9J9C1 A0A1Y1LJL9 H9JIA8 A0A195C7P8 A0A194RAG7 A0A151XF58 A0A2A4JL65 X1WNA5 A0A224XG39 A0A2S2NTG1 X1WSV3 S4NIJ2 A0A154P493 A0A1Y1KV73 J9LIE4 A0A151XET3 A0A023F0J1 A0A151JF33 J9KJ92 A0A2P8XCR5
EMBL
JTDY01003385
KOB69639.1
BABH01003797
BABH01003798
JTDY01001733
KOB73051.1
+ More
GEZM01103137 GEZM01103136 JAV51535.1 GDQN01000847 JAT90207.1 JTDY01001504 KOB73710.1 RSAL01000210 RVE44241.1 GDQN01011628 JAT79426.1 KQ981073 KYN09793.1 GALX01002427 JAB66039.1 GALX01005110 JAB63356.1 KQ977342 KYN03430.1 GEZM01012999 JAV92824.1 KQ982464 KYQ56128.1 KQ977032 KYN06203.1 JTDY01011642 KOB54794.1 KQ977959 KYM98432.1 KQ982064 KYQ60564.1 KQ977963 KYM98425.1 JTDY01001654 KOB73239.1 KQ981002 KYN10407.1 LBMM01018814 KMQ83686.1 ODYU01008608 SOQ52337.1 KQ978178 KYM96586.1 KQ973442 KXZ75531.1 JTDY01003341 KOB69703.1 JTDY01006572 KOB65881.1 ODYU01008829 SOQ52773.1 KQ976954 KYN06860.1 KQ978056 KYM97755.1 JTDY01001855 KOB72712.1 GGMR01015704 MBY28323.1 KQ978855 KYN27942.1 ABLF02022912 ABLF02038819 ABLF02051942 ABLF02060282 KQ978790 KYN28351.1 LBMM01005768 KMQ91221.1 KQ979276 KYN22031.1 KQ460036 KPJ18393.1 KQ981127 KYN09299.1 JTDY01006550 JTDY01002030 JTDY01001260 KOB65898.1 KOB72276.1 KOB74365.1 BABH01004450 GEZM01055231 JAV73081.1 BABH01022219 BABH01022220 KQ978164 KYM96665.1 KQ460500 KPJ14270.1 KQ982204 KYQ59009.1 NWSH01001201 PCG72150.1 ABLF02011452 GFTR01008874 JAW07552.1 GGMR01007417 MBY20036.1 ABLF02028302 ABLF02041631 GAIX01014054 JAA78506.1 KQ434810 KZC06736.1 GEZM01078370 GEZM01078366 GEZM01078363 JAV62757.1 ABLF02034472 ABLF02034478 ABLF02037563 ABLF02037568 ABLF02037575 KQ982228 KYQ58867.1 GBBI01004211 JAC14501.1 KQ979016 KYN24380.1 ABLF02022810 PYGN01002987 PSN29774.1
GEZM01103137 GEZM01103136 JAV51535.1 GDQN01000847 JAT90207.1 JTDY01001504 KOB73710.1 RSAL01000210 RVE44241.1 GDQN01011628 JAT79426.1 KQ981073 KYN09793.1 GALX01002427 JAB66039.1 GALX01005110 JAB63356.1 KQ977342 KYN03430.1 GEZM01012999 JAV92824.1 KQ982464 KYQ56128.1 KQ977032 KYN06203.1 JTDY01011642 KOB54794.1 KQ977959 KYM98432.1 KQ982064 KYQ60564.1 KQ977963 KYM98425.1 JTDY01001654 KOB73239.1 KQ981002 KYN10407.1 LBMM01018814 KMQ83686.1 ODYU01008608 SOQ52337.1 KQ978178 KYM96586.1 KQ973442 KXZ75531.1 JTDY01003341 KOB69703.1 JTDY01006572 KOB65881.1 ODYU01008829 SOQ52773.1 KQ976954 KYN06860.1 KQ978056 KYM97755.1 JTDY01001855 KOB72712.1 GGMR01015704 MBY28323.1 KQ978855 KYN27942.1 ABLF02022912 ABLF02038819 ABLF02051942 ABLF02060282 KQ978790 KYN28351.1 LBMM01005768 KMQ91221.1 KQ979276 KYN22031.1 KQ460036 KPJ18393.1 KQ981127 KYN09299.1 JTDY01006550 JTDY01002030 JTDY01001260 KOB65898.1 KOB72276.1 KOB74365.1 BABH01004450 GEZM01055231 JAV73081.1 BABH01022219 BABH01022220 KQ978164 KYM96665.1 KQ460500 KPJ14270.1 KQ982204 KYQ59009.1 NWSH01001201 PCG72150.1 ABLF02011452 GFTR01008874 JAW07552.1 GGMR01007417 MBY20036.1 ABLF02028302 ABLF02041631 GAIX01014054 JAA78506.1 KQ434810 KZC06736.1 GEZM01078370 GEZM01078366 GEZM01078363 JAV62757.1 ABLF02034472 ABLF02034478 ABLF02037563 ABLF02037568 ABLF02037575 KQ982228 KYQ58867.1 GBBI01004211 JAC14501.1 KQ979016 KYN24380.1 ABLF02022810 PYGN01002987 PSN29774.1
Proteomes
Pfam
Interpro
IPR005312
DUF1759
+ More
IPR040676 DUF5641
IPR008737 Peptidase_asp_put
IPR001969 Aspartic_peptidase_AS
IPR036397 RNaseH_sf
IPR012337 RNaseH-like_sf
IPR008042 Retrotrans_Pao
IPR001584 Integrase_cat-core
IPR021109 Peptidase_aspartic_dom_sf
IPR041588 Integrase_H2C2
IPR001878 Znf_CCHC
IPR001995 Peptidase_A2_cat
IPR008906 HATC_C_dom
IPR040676 DUF5641
IPR008737 Peptidase_asp_put
IPR001969 Aspartic_peptidase_AS
IPR036397 RNaseH_sf
IPR012337 RNaseH-like_sf
IPR008042 Retrotrans_Pao
IPR001584 Integrase_cat-core
IPR021109 Peptidase_aspartic_dom_sf
IPR041588 Integrase_H2C2
IPR001878 Znf_CCHC
IPR001995 Peptidase_A2_cat
IPR008906 HATC_C_dom
Gene 3D
ProteinModelPortal
A0A0L7L2Y3
H9JTH4
A0A0L7LCW9
A0A1Y1JVV0
A0A1E1WTE8
A0A0L7LE91
+ More
A0A3S2P870 A0A1E1VXH0 A0A151ISI5 V5GPJ6 V5I888 A0A151IJK3 A0A1Y1N4T1 A0A151X752 A0A151IMM1 A0A0L7K424 A0A151IDJ5 A0A151XJI2 A0A151IDD4 A0A0L7LCW4 A0A151ITI6 A0A0J7K0K5 A0A2H1WH28 A0A151IAU9 A0A139W8E1 A0A0L7L2J2 A0A0L7KS35 A0A2H1WI74 A0A151IN79 A0A151ICE7 A0A0L7LB62 A0A2S2PFV9 A0A151JNR8 X1XTY9 A0A151JNU0 A0A0J7KLX1 A0A151JAJ1 A0A0N1IHU1 A0A151IRH4 A0A0L7KRI4 H9J9C1 A0A1Y1LJL9 H9JIA8 A0A195C7P8 A0A194RAG7 A0A151XF58 A0A2A4JL65 X1WNA5 A0A224XG39 A0A2S2NTG1 X1WSV3 S4NIJ2 A0A154P493 A0A1Y1KV73 J9LIE4 A0A151XET3 A0A023F0J1 A0A151JF33 J9KJ92 A0A2P8XCR5
A0A3S2P870 A0A1E1VXH0 A0A151ISI5 V5GPJ6 V5I888 A0A151IJK3 A0A1Y1N4T1 A0A151X752 A0A151IMM1 A0A0L7K424 A0A151IDJ5 A0A151XJI2 A0A151IDD4 A0A0L7LCW4 A0A151ITI6 A0A0J7K0K5 A0A2H1WH28 A0A151IAU9 A0A139W8E1 A0A0L7L2J2 A0A0L7KS35 A0A2H1WI74 A0A151IN79 A0A151ICE7 A0A0L7LB62 A0A2S2PFV9 A0A151JNR8 X1XTY9 A0A151JNU0 A0A0J7KLX1 A0A151JAJ1 A0A0N1IHU1 A0A151IRH4 A0A0L7KRI4 H9J9C1 A0A1Y1LJL9 H9JIA8 A0A195C7P8 A0A194RAG7 A0A151XF58 A0A2A4JL65 X1WNA5 A0A224XG39 A0A2S2NTG1 X1WSV3 S4NIJ2 A0A154P493 A0A1Y1KV73 J9LIE4 A0A151XET3 A0A023F0J1 A0A151JF33 J9KJ92 A0A2P8XCR5
Ontologies
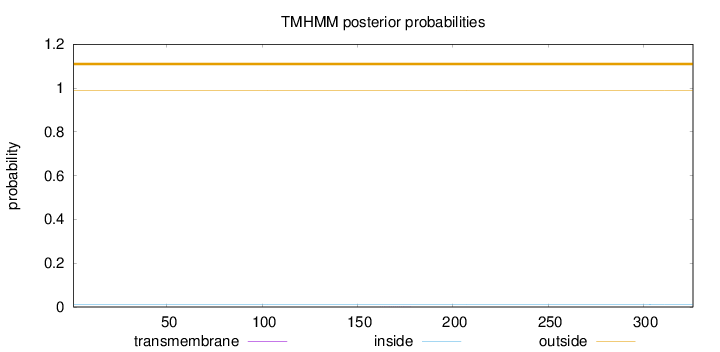
Topology
Length:
326
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00358
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01205
outside
1 - 326
Population Genetic Test Statistics
Pi
318.514444
Theta
178.234919
Tajima's D
2.476175
CLR
0.044815
CSRT
0.943402829858507
Interpretation
Uncertain
