Gene
KWMTBOMO13604
Pre Gene Modal
BGIBMGA011007
Annotation
PREDICTED:_methionine_synthase_reductase_[Papilio_polytes]
Full name
Methionine synthase reductase
+ More
NADPH--cytochrome P450 reductase
NADPH--cytochrome P450 reductase
Location in the cell
Cytoplasmic Reliability : 1.798 Nuclear Reliability : 2.111
Sequence
CDS
ATGGTAGTGATTCAATCTCTAAGAGATACTTTGCAAGCACTACCAGAAAAAACTGTATTTACTTTACCTACATTAACGAAAGTGATTTCTTTGAAACTCGAATACAGAGTGGGAGATGCAAACACAACAATTTATAATGGACCAGACCCAAAATTACCATTTGCTAATTCTGAGCTATTTTATGCTACAATTTCTCAATCAAGGAGGTTGACTGCTTGTGATCAAGGATGCAAAGATGTTTATGAAATCACTTTTGATGTGAAGGATAGTAATTTCACTTTCAAGGCTGGTGATACAATAGGGAATAAAGAAAGTAAAATACCAACACATGTACCTGTTAAATCAACGTTGAGACAAGTGTTGACTGATTGTTTGGATCTTCGGTGTTTGCTTAAAAAGTTGTTCCTGTTAGCGATATCGAGATATACAAAAGAAGATAACGAAAGACAAGTACTCGAATATTTAAGCAGCAAGGAAGGATCTACAGCATATAAAACTCACATATTAAACAAACAAATAAACATTTTGGATATATTCACACTGTTTCCATCCTGCAAGCCACCAATAGAAGTATTATTATCCTATTTACCGAAGCTACTGCCTAGACCATATTCTATAGTCAATTCATATTTAGAAGACAGTAATGCAATTAAAATTTGCTTCTCCGTAATGAATTTAGGAAATAACAGAAGAGGTCTAACAACTGGTTTGTTGGAAGATCACATTTTGAACAAAAACATCAATCTGGAGCATAGTCTAAATGATTTATGTTTAACAGACCATAAACAGAGAAGTGAGTTCAAGGTGCCTATTTATATGAGGAAGAATATTTCCGGGTTCAGTTTGTCTGAAAATTTAGAAACTCCATTGATATTCATTGGTCCCGGTACTGGAGTAGCTCCCTTTATAGGATTTTTACAAGAAATAGAGTATGCAAAGAAGAAGAATTCTAAAGGAAAGTTTGGTACAGTCGACCTGTTTTTCGGATGCAGGAATCCCGAATTAGATTTTATCTACGAGAAGGAACTAAAAGGATTTTTGTCACGGAACGTATTGAATAGTTTGAATGTATCTTTTTCGAGAGTTGATAACAATGAGGTCAAATACGTTCAGGATGCTGTTATGAATGAAGGAGAAAAGATAGCTCAGTTAATAAAAGATGGAGCAAATATATTTGTTTGCGGAGATGTTAGAACGATAGCAGAACAAATAAAAGATGTCTTAATTAAATGCCTTACTAAATATGATAATTGTTTAGAGAATCATGAGAAATATATAGTGCAGATGGAGAAAGACAGAAGATATATTGTGGACGCTTGGTGTTAA
Protein
MVVIQSLRDTLQALPEKTVFTLPTLTKVISLKLEYRVGDANTTIYNGPDPKLPFANSELFYATISQSRRLTACDQGCKDVYEITFDVKDSNFTFKAGDTIGNKESKIPTHVPVKSTLRQVLTDCLDLRCLLKKLFLLAISRYTKEDNERQVLEYLSSKEGSTAYKTHILNKQINILDIFTLFPSCKPPIEVLLSYLPKLLPRPYSIVNSYLEDSNAIKICFSVMNLGNNRRGLTTGLLEDHILNKNINLEHSLNDLCLTDHKQRSEFKVPIYMRKNISGFSLSENLETPLIFIGPGTGVAPFIGFLQEIEYAKKKNSKGKFGTVDLFFGCRNPELDFIYEKELKGFLSRNVLNSLNVSFSRVDNNEVKYVQDAVMNEGEKIAQLIKDGANIFVCGDVRTIAEQIKDVLIKCLTKYDNCLENHEKYIVQMEKDRRYIVDAWC
Summary
Description
Involved in the reductive regeneration of cob(I)alamin (vitamin B12) cofactor required for the maintenance of methionine synthase in a functional state. Necessary for utilization of methylgroups from the folate cycle, thereby affecting transgenerational epigenetic inheritance. Folate pathway donates methyl groups necessary for cellular methylation and affects different pathways such as DNA methylation, possibly explaining the transgenerational epigenetic inheritance effects.
This enzyme is required for electron transfer from NADP to cytochrome P450 in microsomes. It can also provide electron transfer to heme oxygenase and cytochrome B5.
This enzyme is required for electron transfer from NADP to cytochrome P450 in microsomes. It can also provide electron transfer to heme oxygenase and cytochrome B5.
Catalytic Activity
2 [methionine synthase]-methylcob(III)alamin + H(+) + NADP(+) + 2 S-adenosyl-L-homocysteine = 2 [methionine synthase]-cob(II)alamin + NADPH + 2 S-adenosyl-L-methionine
NADPH + 2 oxidized [cytochrome P450] = H(+) + NADP(+) + 2 reduced [cytochrome P450]
NADPH + 2 oxidized [cytochrome P450] = H(+) + NADP(+) + 2 reduced [cytochrome P450]
Cofactor
FAD
FMN
FMN
Biophysicochemical Properties
2.89 uM for NADPH
3540 uM for NADH
3540 uM for NADH
Similarity
Belongs to the AB hydrolase superfamily. Lipase family.
Belongs to the NADPH--cytochrome P450 reductase family.
In the C-terminal section; belongs to the flavoprotein pyridine nucleotide cytochrome reductase family.
In the N-terminal section; belongs to the flavodoxin family.
Belongs to the NADPH--cytochrome P450 reductase family.
In the C-terminal section; belongs to the flavoprotein pyridine nucleotide cytochrome reductase family.
In the N-terminal section; belongs to the flavodoxin family.
Keywords
Alternative splicing
Amino-acid biosynthesis
Complete proteome
Cytoplasm
FAD
Flavoprotein
FMN
Methionine biosynthesis
NADP
Oxidoreductase
Phosphoprotein
Reference proteome
S-adenosyl-L-methionine
3D-structure
Disease mutation
Polymorphism
Feature
chain Methionine synthase reductase
splice variant In isoform 2.
sequence variant Polymorphism; may increase risk for spina bifida; dbSNP:rs1801394.
splice variant In isoform 2.
sequence variant Polymorphism; may increase risk for spina bifida; dbSNP:rs1801394.
Uniprot
H9JNA4
A0A2W1C260
A0A2A4JCR1
A0A2H1WZD3
A0A3S2NMY4
A0A0N0PAV8
+ More
A0A212EJJ6 A0A194PSX0 A0A1W4WUQ3 A0A067R8B5 A0A2J7Q4L9 A0A023F5Y1 Q498R1-2 K1RDM1 A0A0A9ZI51 T1HGG0 Q498R1 A0A210QNW8 A0A1Y1NCC1 A0A162S7X4 H0V865 A0A224XGP0 A0A087UB09 E9H8S9 A0A0P4WEL1 A7S7T0 H0WHQ5 A0A340WK62 A0A0P5VMA6 H2PF60 A0A0P5ZCM3 Q4JIJ2 Q8C1A3 A0A0P5FJJ4 A0A0P5PTH2 A0A2J8TM17 G5AX04 A0A1S3NB54 G3QF79 A0A2I3RPN1 A0A384AWB3 Q9UBK8 Q5RD77 A0A2J8KU04 L8HZY4 A0A3Q7TUF4 S4RH50 F1MGP5 Q9UBK8-1 A0A0P5LRL5 A0A2R9CLM9 H2QQM3 A0A2J8KTZ6 A0A341C0E6 A0A384AWB7 A0A2R9CNP6 F6UGP3 A0A0R4J0G9 A0A2U3V7B0 A0A2Y9I2K6 A0A2Y9SAB4 A0A2Y9P195 A0A158RC93 A0A2K5PVH1 A0A2L2YHN3 A0A3P8ZDH0 A0A1U7UJD2 A0A2L2YFD8 F1PBT9 A0A3B4B5T3 A0A3P8XBR2 A0A1U7QDU5 A0A1S3EXX9 L5M0M4 A0A0P5XZM5 A0A3Q7NYJ3 A0A151ND40 G1SK30 H9F3N1 A0A2K6DWF2 A0A2K5W2G1 A0A2K5W2F3 A0A2K6DWE0 A0A2U3X0A6 A0A2G8KKH1 A0A3M6T547 A0A2K5J5T9 G7MUM6 A0A182K7F5 A0A182XV73 A0A2K5P658 A0A2K6NG07 A0A182RUL9 A0A1D5QVV0 A0A2K6NFZ8 A0A1S3EXX2 A0A2K6LAZ4 S7P4M8 A0A1S3F0G6
A0A212EJJ6 A0A194PSX0 A0A1W4WUQ3 A0A067R8B5 A0A2J7Q4L9 A0A023F5Y1 Q498R1-2 K1RDM1 A0A0A9ZI51 T1HGG0 Q498R1 A0A210QNW8 A0A1Y1NCC1 A0A162S7X4 H0V865 A0A224XGP0 A0A087UB09 E9H8S9 A0A0P4WEL1 A7S7T0 H0WHQ5 A0A340WK62 A0A0P5VMA6 H2PF60 A0A0P5ZCM3 Q4JIJ2 Q8C1A3 A0A0P5FJJ4 A0A0P5PTH2 A0A2J8TM17 G5AX04 A0A1S3NB54 G3QF79 A0A2I3RPN1 A0A384AWB3 Q9UBK8 Q5RD77 A0A2J8KU04 L8HZY4 A0A3Q7TUF4 S4RH50 F1MGP5 Q9UBK8-1 A0A0P5LRL5 A0A2R9CLM9 H2QQM3 A0A2J8KTZ6 A0A341C0E6 A0A384AWB7 A0A2R9CNP6 F6UGP3 A0A0R4J0G9 A0A2U3V7B0 A0A2Y9I2K6 A0A2Y9SAB4 A0A2Y9P195 A0A158RC93 A0A2K5PVH1 A0A2L2YHN3 A0A3P8ZDH0 A0A1U7UJD2 A0A2L2YFD8 F1PBT9 A0A3B4B5T3 A0A3P8XBR2 A0A1U7QDU5 A0A1S3EXX9 L5M0M4 A0A0P5XZM5 A0A3Q7NYJ3 A0A151ND40 G1SK30 H9F3N1 A0A2K6DWF2 A0A2K5W2G1 A0A2K5W2F3 A0A2K6DWE0 A0A2U3X0A6 A0A2G8KKH1 A0A3M6T547 A0A2K5J5T9 G7MUM6 A0A182K7F5 A0A182XV73 A0A2K5P658 A0A2K6NG07 A0A182RUL9 A0A1D5QVV0 A0A2K6NFZ8 A0A1S3EXX2 A0A2K6LAZ4 S7P4M8 A0A1S3F0G6
EC Number
1.16.1.8
1.6.2.4
1.6.2.4
Pubmed
19121390
28756777
26354079
22118469
24845553
25474469
+ More
15489334 22992520 25401762 28812685 28004739 21993624 21292972 17615350 16141072 18413293 21183079 24074862 21993625 22398555 16136131 10564814 9501215 15372022 18221906 20068231 21269460 23186163 24275569 17892308 10484769 10444342 12375236 15979034 22751099 19393038 22722832 17495919 19468303 26561354 25069045 16341006 25463417 22293439 25319552 29023486 30382153 22002653 25244985 25362486 17431167
15489334 22992520 25401762 28812685 28004739 21993624 21292972 17615350 16141072 18413293 21183079 24074862 21993625 22398555 16136131 10564814 9501215 15372022 18221906 20068231 21269460 23186163 24275569 17892308 10484769 10444342 12375236 15979034 22751099 19393038 22722832 17495919 19468303 26561354 25069045 16341006 25463417 22293439 25319552 29023486 30382153 22002653 25244985 25362486 17431167
EMBL
BABH01033329
BABH01033330
BABH01033331
KZ149909
PZC78203.1
NWSH01002102
+ More
PCG69183.1 ODYU01012190 SOQ58367.1 RSAL01000002 RVE55064.1 KQ461153 KPJ08647.1 AGBW02014457 OWR41645.1 KQ459595 KPI95859.1 KK852672 KDR18756.1 NEVH01018378 PNF23516.1 GBBI01002110 JAC16602.1 CH474002 BC100107 JH816847 EKC32196.1 GBHO01001159 JAG42445.1 ACPB03015962 NEDP02002601 OWF50440.1 GEZM01009521 JAV94540.1 LRGB01000084 KZS21080.1 AAKN02042687 GFTR01004770 JAW11656.1 KK119061 KFM74548.1 GL732605 EFX71864.1 GDRN01069008 JAI64109.1 DS469594 EDO40246.1 AAQR03065011 AAQR03065012 AAQR03065013 AAQR03065014 AAQR03065015 AAQR03065016 GDIP01098086 JAM05629.1 ABGA01183291 ABGA01183292 ABGA01183293 NDHI03003490 PNJ34098.1 GDIP01047397 JAM56318.1 DQ084520 AK028628 AK155359 AK163449 CH466563 BC025942 GDIQ01256118 JAJ95606.1 GDIQ01144565 JAL07161.1 PNJ34090.1 JH167284 EHB01565.1 CABD030106542 CABD030106543 AACZ04025692 AF121213 AF121202 AF121203 AF121204 AF121205 AF121206 AF121207 AF121208 AF121209 AF121210 AF121211 AF121212 AF121214 AF025794 AC010346 AC025174 BC054816 BC109216 CR858039 CAH90280.1 NBAG03000338 PNI38500.1 JH882422 ELR49493.1 GDIQ01190608 JAK61117.1 AJFE02028228 PNI38499.1 CT033781 UYYF01004445 VDN04179.1 IAAA01021001 LAA06735.1 IAAA01021002 LAA06737.1 AAEX03017166 KB106364 ELK31263.1 GDIP01066844 JAM36871.1 AKHW03003364 KYO34660.1 JU325484 AFE69240.1 AQIA01057747 AQIA01057748 MRZV01000518 PIK48504.1 RCHS01004322 RMX36468.1 CM001258 EHH26393.1 JSUE03035350 KE161240 EPQ02507.1
PCG69183.1 ODYU01012190 SOQ58367.1 RSAL01000002 RVE55064.1 KQ461153 KPJ08647.1 AGBW02014457 OWR41645.1 KQ459595 KPI95859.1 KK852672 KDR18756.1 NEVH01018378 PNF23516.1 GBBI01002110 JAC16602.1 CH474002 BC100107 JH816847 EKC32196.1 GBHO01001159 JAG42445.1 ACPB03015962 NEDP02002601 OWF50440.1 GEZM01009521 JAV94540.1 LRGB01000084 KZS21080.1 AAKN02042687 GFTR01004770 JAW11656.1 KK119061 KFM74548.1 GL732605 EFX71864.1 GDRN01069008 JAI64109.1 DS469594 EDO40246.1 AAQR03065011 AAQR03065012 AAQR03065013 AAQR03065014 AAQR03065015 AAQR03065016 GDIP01098086 JAM05629.1 ABGA01183291 ABGA01183292 ABGA01183293 NDHI03003490 PNJ34098.1 GDIP01047397 JAM56318.1 DQ084520 AK028628 AK155359 AK163449 CH466563 BC025942 GDIQ01256118 JAJ95606.1 GDIQ01144565 JAL07161.1 PNJ34090.1 JH167284 EHB01565.1 CABD030106542 CABD030106543 AACZ04025692 AF121213 AF121202 AF121203 AF121204 AF121205 AF121206 AF121207 AF121208 AF121209 AF121210 AF121211 AF121212 AF121214 AF025794 AC010346 AC025174 BC054816 BC109216 CR858039 CAH90280.1 NBAG03000338 PNI38500.1 JH882422 ELR49493.1 GDIQ01190608 JAK61117.1 AJFE02028228 PNI38499.1 CT033781 UYYF01004445 VDN04179.1 IAAA01021001 LAA06735.1 IAAA01021002 LAA06737.1 AAEX03017166 KB106364 ELK31263.1 GDIP01066844 JAM36871.1 AKHW03003364 KYO34660.1 JU325484 AFE69240.1 AQIA01057747 AQIA01057748 MRZV01000518 PIK48504.1 RCHS01004322 RMX36468.1 CM001258 EHH26393.1 JSUE03035350 KE161240 EPQ02507.1
Proteomes
UP000005204
UP000218220
UP000283053
UP000053240
UP000007151
UP000053268
+ More
UP000192223 UP000027135 UP000235965 UP000002494 UP000005408 UP000015103 UP000242188 UP000076858 UP000005447 UP000054359 UP000000305 UP000001593 UP000005225 UP000265300 UP000001595 UP000009136 UP000000589 UP000006813 UP000087266 UP000001519 UP000002277 UP000261681 UP000005640 UP000286640 UP000245300 UP000240080 UP000252040 UP000002280 UP000245320 UP000248481 UP000248484 UP000248483 UP000046394 UP000276776 UP000233040 UP000265140 UP000189704 UP000002254 UP000261520 UP000189706 UP000081671 UP000286641 UP000050525 UP000001811 UP000233120 UP000233100 UP000245340 UP000230750 UP000275408 UP000233080 UP000075881 UP000076408 UP000233060 UP000233200 UP000075900 UP000006718 UP000233180
UP000192223 UP000027135 UP000235965 UP000002494 UP000005408 UP000015103 UP000242188 UP000076858 UP000005447 UP000054359 UP000000305 UP000001593 UP000005225 UP000265300 UP000001595 UP000009136 UP000000589 UP000006813 UP000087266 UP000001519 UP000002277 UP000261681 UP000005640 UP000286640 UP000245300 UP000240080 UP000252040 UP000002280 UP000245320 UP000248481 UP000248484 UP000248483 UP000046394 UP000276776 UP000233040 UP000265140 UP000189704 UP000002254 UP000261520 UP000189706 UP000081671 UP000286641 UP000050525 UP000001811 UP000233120 UP000233100 UP000245340 UP000230750 UP000275408 UP000233080 UP000075881 UP000076408 UP000233060 UP000233200 UP000075900 UP000006718 UP000233180
Interpro
IPR017938
Riboflavin_synthase-like_b-brl
+ More
IPR017927 FAD-bd_FR_type
IPR001433 OxRdtase_FAD/NAD-bd
IPR001709 Flavoprot_Pyr_Nucl_cyt_Rdtase
IPR023173 NADPH_Cyt_P450_Rdtase_alpha
IPR003097 CysJ-like_FAD-binding
IPR039261 FNR_nucleotide-bd
IPR033906 Lipase_N
IPR013818 Lipase/vitellogenin
IPR029058 AB_hydrolase
IPR029039 Flavoprotein-like_sf
IPR001094 Flavdoxin-like
IPR008254 Flavodoxin/NO_synth
IPR023208 P450R
IPR017927 FAD-bd_FR_type
IPR001433 OxRdtase_FAD/NAD-bd
IPR001709 Flavoprot_Pyr_Nucl_cyt_Rdtase
IPR023173 NADPH_Cyt_P450_Rdtase_alpha
IPR003097 CysJ-like_FAD-binding
IPR039261 FNR_nucleotide-bd
IPR033906 Lipase_N
IPR013818 Lipase/vitellogenin
IPR029058 AB_hydrolase
IPR029039 Flavoprotein-like_sf
IPR001094 Flavdoxin-like
IPR008254 Flavodoxin/NO_synth
IPR023208 P450R
Gene 3D
ProteinModelPortal
H9JNA4
A0A2W1C260
A0A2A4JCR1
A0A2H1WZD3
A0A3S2NMY4
A0A0N0PAV8
+ More
A0A212EJJ6 A0A194PSX0 A0A1W4WUQ3 A0A067R8B5 A0A2J7Q4L9 A0A023F5Y1 Q498R1-2 K1RDM1 A0A0A9ZI51 T1HGG0 Q498R1 A0A210QNW8 A0A1Y1NCC1 A0A162S7X4 H0V865 A0A224XGP0 A0A087UB09 E9H8S9 A0A0P4WEL1 A7S7T0 H0WHQ5 A0A340WK62 A0A0P5VMA6 H2PF60 A0A0P5ZCM3 Q4JIJ2 Q8C1A3 A0A0P5FJJ4 A0A0P5PTH2 A0A2J8TM17 G5AX04 A0A1S3NB54 G3QF79 A0A2I3RPN1 A0A384AWB3 Q9UBK8 Q5RD77 A0A2J8KU04 L8HZY4 A0A3Q7TUF4 S4RH50 F1MGP5 Q9UBK8-1 A0A0P5LRL5 A0A2R9CLM9 H2QQM3 A0A2J8KTZ6 A0A341C0E6 A0A384AWB7 A0A2R9CNP6 F6UGP3 A0A0R4J0G9 A0A2U3V7B0 A0A2Y9I2K6 A0A2Y9SAB4 A0A2Y9P195 A0A158RC93 A0A2K5PVH1 A0A2L2YHN3 A0A3P8ZDH0 A0A1U7UJD2 A0A2L2YFD8 F1PBT9 A0A3B4B5T3 A0A3P8XBR2 A0A1U7QDU5 A0A1S3EXX9 L5M0M4 A0A0P5XZM5 A0A3Q7NYJ3 A0A151ND40 G1SK30 H9F3N1 A0A2K6DWF2 A0A2K5W2G1 A0A2K5W2F3 A0A2K6DWE0 A0A2U3X0A6 A0A2G8KKH1 A0A3M6T547 A0A2K5J5T9 G7MUM6 A0A182K7F5 A0A182XV73 A0A2K5P658 A0A2K6NG07 A0A182RUL9 A0A1D5QVV0 A0A2K6NFZ8 A0A1S3EXX2 A0A2K6LAZ4 S7P4M8 A0A1S3F0G6
A0A212EJJ6 A0A194PSX0 A0A1W4WUQ3 A0A067R8B5 A0A2J7Q4L9 A0A023F5Y1 Q498R1-2 K1RDM1 A0A0A9ZI51 T1HGG0 Q498R1 A0A210QNW8 A0A1Y1NCC1 A0A162S7X4 H0V865 A0A224XGP0 A0A087UB09 E9H8S9 A0A0P4WEL1 A7S7T0 H0WHQ5 A0A340WK62 A0A0P5VMA6 H2PF60 A0A0P5ZCM3 Q4JIJ2 Q8C1A3 A0A0P5FJJ4 A0A0P5PTH2 A0A2J8TM17 G5AX04 A0A1S3NB54 G3QF79 A0A2I3RPN1 A0A384AWB3 Q9UBK8 Q5RD77 A0A2J8KU04 L8HZY4 A0A3Q7TUF4 S4RH50 F1MGP5 Q9UBK8-1 A0A0P5LRL5 A0A2R9CLM9 H2QQM3 A0A2J8KTZ6 A0A341C0E6 A0A384AWB7 A0A2R9CNP6 F6UGP3 A0A0R4J0G9 A0A2U3V7B0 A0A2Y9I2K6 A0A2Y9SAB4 A0A2Y9P195 A0A158RC93 A0A2K5PVH1 A0A2L2YHN3 A0A3P8ZDH0 A0A1U7UJD2 A0A2L2YFD8 F1PBT9 A0A3B4B5T3 A0A3P8XBR2 A0A1U7QDU5 A0A1S3EXX9 L5M0M4 A0A0P5XZM5 A0A3Q7NYJ3 A0A151ND40 G1SK30 H9F3N1 A0A2K6DWF2 A0A2K5W2G1 A0A2K5W2F3 A0A2K6DWE0 A0A2U3X0A6 A0A2G8KKH1 A0A3M6T547 A0A2K5J5T9 G7MUM6 A0A182K7F5 A0A182XV73 A0A2K5P658 A0A2K6NG07 A0A182RUL9 A0A1D5QVV0 A0A2K6NFZ8 A0A1S3EXX2 A0A2K6LAZ4 S7P4M8 A0A1S3F0G6
PDB
2QTZ
E-value=3.19443e-50,
Score=502
Ontologies
GO
GO:0016491
GO:0005576
GO:0006306
GO:0016723
GO:0005829
GO:0009086
GO:0050667
GO:0010181
GO:0003958
GO:0071949
GO:0050444
GO:0050660
GO:0046655
GO:1904042
GO:0043418
GO:0030586
GO:0070402
GO:0033353
GO:0055114
GO:0047138
GO:0050821
GO:0006555
GO:0009235
GO:0000096
GO:0050661
GO:0032259
GO:0016021
GO:0005789
GO:0006810
GO:0006520
GO:0006508
GO:0008237
GO:0008152
GO:0016787
GO:0003676
GO:0015930
Topology
Subcellular location
Secreted
Cytoplasm
Endoplasmic reticulum membrane
Cytoplasm
Endoplasmic reticulum membrane
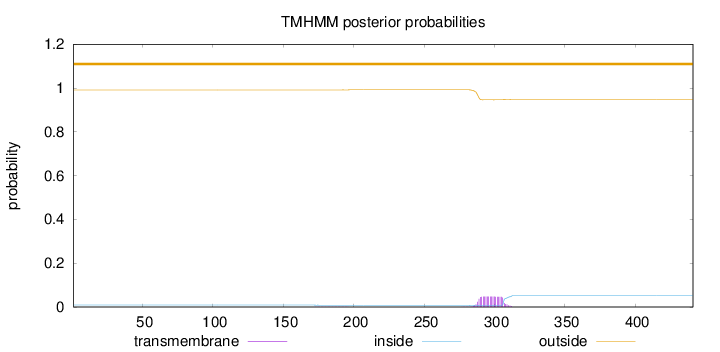
Length:
441
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.95043
Exp number, first 60 AAs:
0.00042
Total prob of N-in:
0.00865
outside
1 - 441
Population Genetic Test Statistics
Pi
319.984897
Theta
195.387804
Tajima's D
1.910458
CLR
0.164169
CSRT
0.866556672166392
Interpretation
Uncertain
