Pre Gene Modal
BGIBMGA011003
Annotation
PREDICTED:_UDP-N-acetylglucosamine_transferase_subunit_ALG13_homolog_[Plutella_xylostella]
Full name
Putative bifunctional UDP-N-acetylglucosamine transferase and deubiquitinase ALG13
Alternative Name
Asparagine-linked glycosylation 13 homolog
Glycosyltransferase 28 domain-containing protein 1
UDP-N-acetylglucosamine transferase subunit ALG13 homolog
Glycosyltransferase 28 domain-containing protein 1
UDP-N-acetylglucosamine transferase subunit ALG13 homolog
Location in the cell
Cytoplasmic Reliability : 3.179
Sequence
CDS
ATGCTTTCACCAGCCGTAATTCATGCACTAAAGGAGATCGGCTGCAAAGAGGTTGTAATACAAATTGGCAATAGTGACGTAGATCCAGGAAATTTTACTAAAAACGAGATTAAGTTCACTTTGTACAGATTTAAGGACTCTATATTAGAAGATATAAAAAAAGCCGATTTAGTTATAAGTCACGCCGGAGCAGGAAGCTGTCTTGAAGTGTTAGAAGAAAAGAAACCTCTTTTAGTGGTTGTTAACGAGGATTTGATGGACAACCACCAACTAGAACTCGCAGAACAGCTCCAAATAGATGGACATTTATACTATTGTACTTGCGATACTCTGGTCAACACACTAAATGTTGTTGACTTTGAATTATTACGTCCCTTTCCCAAAGCAAATCCTATTTTGTTTGTCAGATATTTGGATGATATTTTCAGAGTATATAAAAATATAGACTGA
Protein
MLSPAVIHALKEIGCKEVVIQIGNSDVDPGNFTKNEIKFTLYRFKDSILEDIKKADLVISHAGAGSCLEVLEEKKPLLVVVNEDLMDNHQLELAEQLQIDGHLYYCTCDTLVNTLNVVDFELLRPFPKANPILFVRYLDDIFRVYKNID
Summary
Catalytic Activity
N-acetyl-alpha-D-glucosaminyl-diphosphodolichol + UDP-N-acetyl-alpha-D-glucosamine = H(+) + N,N'-diacetylchitobiosyl diphosphodolichol + UDP
Thiol-dependent hydrolysis of ester, thioester, amide, peptide and isopeptide bonds formed by the C-terminal Gly of ubiquitin (a 76-residue protein attached to proteins as an intracellular targeting signal).
Thiol-dependent hydrolysis of ester, thioester, amide, peptide and isopeptide bonds formed by the C-terminal Gly of ubiquitin (a 76-residue protein attached to proteins as an intracellular targeting signal).
Subunit
Isoform 2 may interact with ALG14.
Similarity
Belongs to the glycosyltransferase 28 family.
Keywords
Alternative splicing
Complete proteome
Congenital disorder of glycosylation
Disease mutation
Endoplasmic reticulum
Epilepsy
Glycosyltransferase
Hydrolase
Multifunctional enzyme
Polymorphism
Protease
Reference proteome
Thiol protease
Transferase
Ubl conjugation pathway
Feature
chain Putative bifunctional UDP-N-acetylglucosamine transferase and deubiquitinase ALG13
splice variant In isoform 4.
sequence variant In EIEE36; disease features include abnormal isoelectric focusing of serum transferrin consistent with a glycosylation defect; enzyme activity at about 17% of wild-type; dbSNP:rs867599353.
splice variant In isoform 4.
sequence variant In EIEE36; disease features include abnormal isoelectric focusing of serum transferrin consistent with a glycosylation defect; enzyme activity at about 17% of wild-type; dbSNP:rs867599353.
Uniprot
H9JNA0
A0A2W1BX24
A0A2H1WXA9
A0A0L7LHT1
A0A0N1I546
A0A3S2TTT7
+ More
S4PL85 V9LJ87 F7BAW9 A0A151PDS9 A0A3Q0RTP5 K1RHC5 A0A2D4KJU8 A0A2D4PJ01 F7E7G8 A0A2P4TIL5 A0A1D5PWJ2 A0A3Q3ESM9 A0A226NJC2 A0A226P6D7 A0A3B4G942 A0A3Q2VT39 A0A3Q4GBN7 I3JD92 A0A3P9JIF7 A0A3B3HEY0 Q4RP99 A0A3Q3EP66 A0A3P9LDW7 A0A0K8RYE1 A0A3S2MUV2 A0A3P9BA76 A0A3P8QC11 A0A3B3CER7 A0A3B5KPX6 A0A2I4AZX6 A0A1L1RT72 A0A3P8X979 A0A2I0UAA5 A0A3Q2TDJ1 A0A146P5A6 A0A1A8JYN3 A0A1A8EJN9 V8NLS8 A0A3B4AZ89 A0A1A8SGB6 A0A1V4KYP4 A0A1A8N2Q8 A0A1A8BTL9 A0A1A7ZBL4 G3PXZ1 A0A3Q3VNA0 A0A3Q2DAF6 A0A2U9C8I5 A0A2I3MNS4 A0A3Q2ECG1 A0A287B9F4 A0A1A7X791 A0A3P8V6M2 A0A3Q2ZNN7 A0A341BQJ4 A0A3B3WYA9 A0A3Q1IID4 U3FXV1 A0A1W4YLS7 A0A2K5DDH0 W5NCI7 A0A2K6SFU3 A0A2J8VAF9 A0A2Y9DRL6 S7NRJ4 A0A2K5PW92 F6YIZ0 A0A2K5JKH5 A0A2R9CST1 H2QZ05 A0A2K6CHW9 A0A2K5VHX9 F7CXN6 Q9NP73-2 A0A2K5ZYT9 A0A2K5LYR0 A0A2I3G9S4 E2RPL3 A0A087XS90 A0A2I2U8L0 M1ECH4 M4AF83 A0A3B5MIM4 A0A315VHN8 K9IYD3 F6PT67 A0A1U7R7C4 A0A061HXS5 A0A3B3U2T6 A0A3Q0CGD1 A0A2K6PVW6 A0A3P9NTD5 A0A0A9ZGQ7 A0A3S3Q999
S4PL85 V9LJ87 F7BAW9 A0A151PDS9 A0A3Q0RTP5 K1RHC5 A0A2D4KJU8 A0A2D4PJ01 F7E7G8 A0A2P4TIL5 A0A1D5PWJ2 A0A3Q3ESM9 A0A226NJC2 A0A226P6D7 A0A3B4G942 A0A3Q2VT39 A0A3Q4GBN7 I3JD92 A0A3P9JIF7 A0A3B3HEY0 Q4RP99 A0A3Q3EP66 A0A3P9LDW7 A0A0K8RYE1 A0A3S2MUV2 A0A3P9BA76 A0A3P8QC11 A0A3B3CER7 A0A3B5KPX6 A0A2I4AZX6 A0A1L1RT72 A0A3P8X979 A0A2I0UAA5 A0A3Q2TDJ1 A0A146P5A6 A0A1A8JYN3 A0A1A8EJN9 V8NLS8 A0A3B4AZ89 A0A1A8SGB6 A0A1V4KYP4 A0A1A8N2Q8 A0A1A8BTL9 A0A1A7ZBL4 G3PXZ1 A0A3Q3VNA0 A0A3Q2DAF6 A0A2U9C8I5 A0A2I3MNS4 A0A3Q2ECG1 A0A287B9F4 A0A1A7X791 A0A3P8V6M2 A0A3Q2ZNN7 A0A341BQJ4 A0A3B3WYA9 A0A3Q1IID4 U3FXV1 A0A1W4YLS7 A0A2K5DDH0 W5NCI7 A0A2K6SFU3 A0A2J8VAF9 A0A2Y9DRL6 S7NRJ4 A0A2K5PW92 F6YIZ0 A0A2K5JKH5 A0A2R9CST1 H2QZ05 A0A2K6CHW9 A0A2K5VHX9 F7CXN6 Q9NP73-2 A0A2K5ZYT9 A0A2K5LYR0 A0A2I3G9S4 E2RPL3 A0A087XS90 A0A2I2U8L0 M1ECH4 M4AF83 A0A3B5MIM4 A0A315VHN8 K9IYD3 F6PT67 A0A1U7R7C4 A0A061HXS5 A0A3B3U2T6 A0A3Q0CGD1 A0A2K6PVW6 A0A3P9NTD5 A0A0A9ZGQ7 A0A3S3Q999
Pubmed
19121390
28756777
26227816
26354079
23622113
24402279
+ More
18464734 22293439 22992520 17495919 15592404 24621616 25186727 17554307 15496914 25727380 29451363 21551351 25069045 24297900 25463417 30723633 24487278 25243066 22722832 16136131 17431167 22002653 25319552 14702039 15772651 15489334 16100110 23827681 21269460 22492991 23934111 26138355 23033978 27476654 16341006 17975172 23236062 23542700 29703783 19892987 23929341 29704459 28071753 25362486 25401762
18464734 22293439 22992520 17495919 15592404 24621616 25186727 17554307 15496914 25727380 29451363 21551351 25069045 24297900 25463417 30723633 24487278 25243066 22722832 16136131 17431167 22002653 25319552 14702039 15772651 15489334 16100110 23827681 21269460 22492991 23934111 26138355 23033978 27476654 16341006 17975172 23236062 23542700 29703783 19892987 23929341 29704459 28071753 25362486 25401762
EMBL
BABH01033318
KZ149909
PZC78195.1
ODYU01011769
SOQ57700.1
JTDY01001053
+ More
KOB75007.1 KQ461153 KPJ08641.1 RSAL01000002 RVE55006.1 RVE55059.1 GAIX01004175 JAA88385.1 JW880764 AFP13281.1 AAPN01289039 AAPN01289040 AAPN01289041 AKHW03000474 KYO47267.1 JH819176 EKC33531.1 IACL01071176 LAB08968.1 IACN01075325 IACN01075326 LAB57458.1 PPHD01000017 POI36184.1 AADN05000020 MCFN01000034 OXB67624.1 AWGT02000146 OXB75411.1 AERX01012441 AERX01012442 CAAE01015008 CAG09783.1 GBKC01000176 JAG45894.1 CM012446 RVE67368.1 KZ505944 PKU42980.1 GCES01147209 JAQ39113.1 HAED01014544 HAEE01005042 SBR25062.1 HAEB01000570 HAEC01015341 SBQ47022.1 AZIM01002841 ETE63249.1 HAEH01014405 HAEI01014912 SBS17381.1 LSYS01001150 OPJ89401.1 HAEF01003022 HAEG01000169 SBR63373.1 HADZ01006608 HAEA01013032 SBP70549.1 HADY01001221 HAEJ01018750 SBP39706.1 CP026255 AWP11939.1 AHZZ02038378 AEMK02000121 DQIR01257397 HDC12875.1 HADW01012526 HADX01015210 SBP13926.1 GAMP01001430 JAB51325.1 AHAT01016800 NDHI03003425 PNJ54500.1 KE164818 EPQ20339.1 GAMT01000278 GAMS01002024 GAMR01004478 GAMQ01003576 JAB11583.1 JAB21112.1 JAB29454.1 JAB38275.1 AJFE02076159 AJFE02076160 AC193880 GABC01004694 GABF01005266 GABD01009523 GABD01009522 GABE01004809 NBAG03000329 JAA06644.1 JAA16879.1 JAA23577.1 JAA39930.1 PNI40053.1 AQIA01086487 JSUE03046497 JSUE03046498 JU336437 JU470726 CM001273 AFE80190.1 AFH27530.1 EHH31003.1 AF220051 AK026671 AK300394 AK302729 AK302890 AK304712 AK316306 AK316522 AK312229 AL049563 AL096764 CH471120 BC005336 BC117377 BC117379 AK223154 ADFV01008300 ADFV01008301 ADFV01008302 AAEX03026776 AYCK01006064 AANG04001034 JP005447 AER94044.1 NHOQ01002284 PWA18656.1 GABZ01008291 JAA45234.1 KE685696 RAZU01000333 ERE65171.1 RLQ59675.1 GBHO01002395 GBHO01002394 JAG41209.1 JAG41210.1 NCKU01001935 NCKU01000276 RWS10865.1 RWS16183.1
KOB75007.1 KQ461153 KPJ08641.1 RSAL01000002 RVE55006.1 RVE55059.1 GAIX01004175 JAA88385.1 JW880764 AFP13281.1 AAPN01289039 AAPN01289040 AAPN01289041 AKHW03000474 KYO47267.1 JH819176 EKC33531.1 IACL01071176 LAB08968.1 IACN01075325 IACN01075326 LAB57458.1 PPHD01000017 POI36184.1 AADN05000020 MCFN01000034 OXB67624.1 AWGT02000146 OXB75411.1 AERX01012441 AERX01012442 CAAE01015008 CAG09783.1 GBKC01000176 JAG45894.1 CM012446 RVE67368.1 KZ505944 PKU42980.1 GCES01147209 JAQ39113.1 HAED01014544 HAEE01005042 SBR25062.1 HAEB01000570 HAEC01015341 SBQ47022.1 AZIM01002841 ETE63249.1 HAEH01014405 HAEI01014912 SBS17381.1 LSYS01001150 OPJ89401.1 HAEF01003022 HAEG01000169 SBR63373.1 HADZ01006608 HAEA01013032 SBP70549.1 HADY01001221 HAEJ01018750 SBP39706.1 CP026255 AWP11939.1 AHZZ02038378 AEMK02000121 DQIR01257397 HDC12875.1 HADW01012526 HADX01015210 SBP13926.1 GAMP01001430 JAB51325.1 AHAT01016800 NDHI03003425 PNJ54500.1 KE164818 EPQ20339.1 GAMT01000278 GAMS01002024 GAMR01004478 GAMQ01003576 JAB11583.1 JAB21112.1 JAB29454.1 JAB38275.1 AJFE02076159 AJFE02076160 AC193880 GABC01004694 GABF01005266 GABD01009523 GABD01009522 GABE01004809 NBAG03000329 JAA06644.1 JAA16879.1 JAA23577.1 JAA39930.1 PNI40053.1 AQIA01086487 JSUE03046497 JSUE03046498 JU336437 JU470726 CM001273 AFE80190.1 AFH27530.1 EHH31003.1 AF220051 AK026671 AK300394 AK302729 AK302890 AK304712 AK316306 AK316522 AK312229 AL049563 AL096764 CH471120 BC005336 BC117377 BC117379 AK223154 ADFV01008300 ADFV01008301 ADFV01008302 AAEX03026776 AYCK01006064 AANG04001034 JP005447 AER94044.1 NHOQ01002284 PWA18656.1 GABZ01008291 JAA45234.1 KE685696 RAZU01000333 ERE65171.1 RLQ59675.1 GBHO01002395 GBHO01002394 JAG41209.1 JAG41210.1 NCKU01001935 NCKU01000276 RWS10865.1 RWS16183.1
Proteomes
UP000005204
UP000037510
UP000053240
UP000283053
UP000002279
UP000050525
+ More
UP000261340 UP000005408 UP000002280 UP000000539 UP000261660 UP000198323 UP000198419 UP000261460 UP000264840 UP000261580 UP000005207 UP000265200 UP000001038 UP000007303 UP000265180 UP000265160 UP000265100 UP000261560 UP000005226 UP000192220 UP000265140 UP000265000 UP000261520 UP000190648 UP000007635 UP000261620 UP000265020 UP000246464 UP000028761 UP000008227 UP000265120 UP000264800 UP000252040 UP000261480 UP000265040 UP000192224 UP000233020 UP000018468 UP000233220 UP000248480 UP000233040 UP000008225 UP000233080 UP000240080 UP000002277 UP000233120 UP000233100 UP000006718 UP000005640 UP000233140 UP000233060 UP000001073 UP000002254 UP000028760 UP000011712 UP000002852 UP000261380 UP000002281 UP000189706 UP000030759 UP000273346 UP000261500 UP000233200 UP000242638 UP000285301
UP000261340 UP000005408 UP000002280 UP000000539 UP000261660 UP000198323 UP000198419 UP000261460 UP000264840 UP000261580 UP000005207 UP000265200 UP000001038 UP000007303 UP000265180 UP000265160 UP000265100 UP000261560 UP000005226 UP000192220 UP000265140 UP000265000 UP000261520 UP000190648 UP000007635 UP000261620 UP000265020 UP000246464 UP000028761 UP000008227 UP000265120 UP000264800 UP000252040 UP000261480 UP000265040 UP000192224 UP000233020 UP000018468 UP000233220 UP000248480 UP000233040 UP000008225 UP000233080 UP000240080 UP000002277 UP000233120 UP000233100 UP000006718 UP000005640 UP000233140 UP000233060 UP000001073 UP000002254 UP000028760 UP000011712 UP000002852 UP000261380 UP000002281 UP000189706 UP000030759 UP000273346 UP000261500 UP000233200 UP000242638 UP000285301
Interpro
SUPFAM
SSF54001
SSF54001
ProteinModelPortal
H9JNA0
A0A2W1BX24
A0A2H1WXA9
A0A0L7LHT1
A0A0N1I546
A0A3S2TTT7
+ More
S4PL85 V9LJ87 F7BAW9 A0A151PDS9 A0A3Q0RTP5 K1RHC5 A0A2D4KJU8 A0A2D4PJ01 F7E7G8 A0A2P4TIL5 A0A1D5PWJ2 A0A3Q3ESM9 A0A226NJC2 A0A226P6D7 A0A3B4G942 A0A3Q2VT39 A0A3Q4GBN7 I3JD92 A0A3P9JIF7 A0A3B3HEY0 Q4RP99 A0A3Q3EP66 A0A3P9LDW7 A0A0K8RYE1 A0A3S2MUV2 A0A3P9BA76 A0A3P8QC11 A0A3B3CER7 A0A3B5KPX6 A0A2I4AZX6 A0A1L1RT72 A0A3P8X979 A0A2I0UAA5 A0A3Q2TDJ1 A0A146P5A6 A0A1A8JYN3 A0A1A8EJN9 V8NLS8 A0A3B4AZ89 A0A1A8SGB6 A0A1V4KYP4 A0A1A8N2Q8 A0A1A8BTL9 A0A1A7ZBL4 G3PXZ1 A0A3Q3VNA0 A0A3Q2DAF6 A0A2U9C8I5 A0A2I3MNS4 A0A3Q2ECG1 A0A287B9F4 A0A1A7X791 A0A3P8V6M2 A0A3Q2ZNN7 A0A341BQJ4 A0A3B3WYA9 A0A3Q1IID4 U3FXV1 A0A1W4YLS7 A0A2K5DDH0 W5NCI7 A0A2K6SFU3 A0A2J8VAF9 A0A2Y9DRL6 S7NRJ4 A0A2K5PW92 F6YIZ0 A0A2K5JKH5 A0A2R9CST1 H2QZ05 A0A2K6CHW9 A0A2K5VHX9 F7CXN6 Q9NP73-2 A0A2K5ZYT9 A0A2K5LYR0 A0A2I3G9S4 E2RPL3 A0A087XS90 A0A2I2U8L0 M1ECH4 M4AF83 A0A3B5MIM4 A0A315VHN8 K9IYD3 F6PT67 A0A1U7R7C4 A0A061HXS5 A0A3B3U2T6 A0A3Q0CGD1 A0A2K6PVW6 A0A3P9NTD5 A0A0A9ZGQ7 A0A3S3Q999
S4PL85 V9LJ87 F7BAW9 A0A151PDS9 A0A3Q0RTP5 K1RHC5 A0A2D4KJU8 A0A2D4PJ01 F7E7G8 A0A2P4TIL5 A0A1D5PWJ2 A0A3Q3ESM9 A0A226NJC2 A0A226P6D7 A0A3B4G942 A0A3Q2VT39 A0A3Q4GBN7 I3JD92 A0A3P9JIF7 A0A3B3HEY0 Q4RP99 A0A3Q3EP66 A0A3P9LDW7 A0A0K8RYE1 A0A3S2MUV2 A0A3P9BA76 A0A3P8QC11 A0A3B3CER7 A0A3B5KPX6 A0A2I4AZX6 A0A1L1RT72 A0A3P8X979 A0A2I0UAA5 A0A3Q2TDJ1 A0A146P5A6 A0A1A8JYN3 A0A1A8EJN9 V8NLS8 A0A3B4AZ89 A0A1A8SGB6 A0A1V4KYP4 A0A1A8N2Q8 A0A1A8BTL9 A0A1A7ZBL4 G3PXZ1 A0A3Q3VNA0 A0A3Q2DAF6 A0A2U9C8I5 A0A2I3MNS4 A0A3Q2ECG1 A0A287B9F4 A0A1A7X791 A0A3P8V6M2 A0A3Q2ZNN7 A0A341BQJ4 A0A3B3WYA9 A0A3Q1IID4 U3FXV1 A0A1W4YLS7 A0A2K5DDH0 W5NCI7 A0A2K6SFU3 A0A2J8VAF9 A0A2Y9DRL6 S7NRJ4 A0A2K5PW92 F6YIZ0 A0A2K5JKH5 A0A2R9CST1 H2QZ05 A0A2K6CHW9 A0A2K5VHX9 F7CXN6 Q9NP73-2 A0A2K5ZYT9 A0A2K5LYR0 A0A2I3G9S4 E2RPL3 A0A087XS90 A0A2I2U8L0 M1ECH4 M4AF83 A0A3B5MIM4 A0A315VHN8 K9IYD3 F6PT67 A0A1U7R7C4 A0A061HXS5 A0A3B3U2T6 A0A3Q0CGD1 A0A2K6PVW6 A0A3P9NTD5 A0A0A9ZGQ7 A0A3S3Q999
PDB
2JZC
E-value=4.77609e-06,
Score=114
Ontologies
PATHWAY
GO
PANTHER
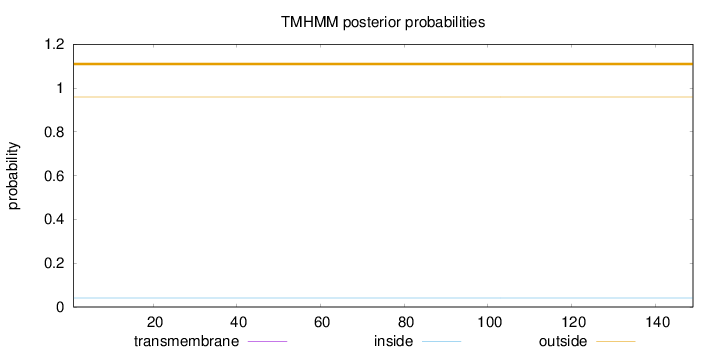
Topology
Subcellular location
Endoplasmic reticulum
Length:
149
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.000900000000000001
Exp number, first 60 AAs:
0
Total prob of N-in:
0.04069
outside
1 - 149
Population Genetic Test Statistics
Pi
165.615708
Theta
187.059252
Tajima's D
-0.360734
CLR
0.434092
CSRT
0.273036348182591
Interpretation
Uncertain
