Gene
KWMTBOMO13595
Pre Gene Modal
BGIBMGA012804
Annotation
PREDICTED:_nucleic-acid-binding_protein_from_mobile_element_jockey-like_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 3.504
Sequence
CDS
ATGAAGTTAAACCCCTTAAAAAACCCTCAACCACAACACAAACAGCTTACTCAACAAGCACTCCTTCCAGGCTTGTTTGACAGACAACGTTCGTCATCTCTTTCTGACCTGGGAAGGTCACCACAAGACACGACCAATGTAGAGCAACCAAATGTAGCCAGTATCGCACCAGTAGCAACCTTGCTGGAAGATATTCCTCAACCCCCTGCTCCTCCACCATGGCAAAGGGTACAATCACCCAATAGCAGAAGCAGTAAGAAAAGAAAGTACTCCAGTTCCTCACCAGAGGCCCCAAGTACCACCGTGAAAAACCGCTTCAGCACCTTACCTTTAGACGCAACGGATGAACCATGTACTACCGACCGCCCTAAACCCAGCAAACCTCCACCTATAGTCCTCAATGGTATAGAAGATGTTACAGAGCTATCGAAGCTCCTTCAAACCGTCAGCAGCAAAGATGATTTCAAATTTAAACTAATTAATAAAAACCTACTCCATATTCTGGTTAACAATGCTGAGGAGTATAAGAAAATCATAGCGGTAATAAGAGCAAATGGACTCATAGGACACACATTCACCCCTAAAGACATCAAATGCTATCGCATTGTCATAAAAGACCTCCACCACAGCACACCCCATGAAGCGATAACAGAAGCCATTGAGGCAACTAACAATAAAATTAAAGGCGAAATCATCAATGCACGTTACGGACCCGACAAGAAACCTACATCTACTTTCTTTGTAAACATTGAGCCTAGTGTAAATAACCCTGCAGCCAAGAATATAACTCATATATTTAACCAAAAGGTCAAGATAGAGGATCCTCGCAAATCAAAAAGTATTGTTCAATGTCAACGTTGTCAGCGCTACGGTCACTCGAAAAATAACTGTATGAGACCATATAGGTGCGTTAAATGCGGTGAGGGTCACAAAACCTCGGAGTGCCAAAAGAAAGATAGAACCACACCAGCTAAATGCGCTCTCTGCTTGTGTGATCACCCGGCTAACTACAAAGGCTGCGAAATTTACAAGGAAATCCTTGCCCGAAGAAACAAAAACTTTACTCCAAGAGAAAGACAACCCGCTCTAGCTATCACAGCCTTGCCCATTAATCCAATCAGCTCCATATCGACAAAAACTGAAGGTATCGAAAGACCAACTCACCAACGAACTTATGCTGAAGTCGCTGATGGAAGGAATTCCCAATCTAATCAACAATCCTACAACAAACTGGAAAACATCCTACTTAAACAAAGTGAGAAAATGGACCTCCTATTACAACAAATAGGCTCCTTAGTGAGTCTTATTACTTCTCTCGTCTCCAAGCTTCTAAAATGA
Protein
MKLNPLKNPQPQHKQLTQQALLPGLFDRQRSSSLSDLGRSPQDTTNVEQPNVASIAPVATLLEDIPQPPAPPPWQRVQSPNSRSSKKRKYSSSSPEAPSTTVKNRFSTLPLDATDEPCTTDRPKPSKPPPIVLNGIEDVTELSKLLQTVSSKDDFKFKLINKNLLHILVNNAEEYKKIIAVIRANGLIGHTFTPKDIKCYRIVIKDLHHSTPHEAITEAIEATNNKIKGEIINARYGPDKKPTSTFFVNIEPSVNNPAAKNITHIFNQKVKIEDPRKSKSIVQCQRCQRYGHSKNNCMRPYRCVKCGEGHKTSECQKKDRTTPAKCALCLCDHPANYKGCEIYKEILARRNKNFTPRERQPALAITALPINPISSISTKTEGIERPTHQRTYAEVADGRNSQSNQQSYNKLENILLKQSEKMDLLLQQIGSLVSLITSLVSKLLK
Summary
Uniprot
H9JTE2
A0A194RK46
A0A2J7QGW8
A0A2J7QDG6
A0A1B0CK87
A0A0J7KKG5
+ More
A0A2A4J8Z7 A0A2S2R608 A0A1Y1N079 J9KE07 A0A1B0FV55 A0A1B0GHS3 A0A1B0CBY8 A0A1B0CMQ7 A0A2S2PDP3 A0A2S2P6J9 A0A2S2NWV3 X1X7V8 X1X2F5 J9LNE5 A0A1B0DHQ9 X1WQY4 J9LJG0 A0A0J7KTN8 A0A2S2NQE0 A0A2S2Q8X5 A0A224XT32 X1WK76 A0A2S2NXZ1 J9LNL0 X1X088 A0A1B0CRN6 A0A2S2R8S1 A0A2S2QG05 J9L3V2 A0A0J7MYU4 A0A2S2P9D3
A0A2A4J8Z7 A0A2S2R608 A0A1Y1N079 J9KE07 A0A1B0FV55 A0A1B0GHS3 A0A1B0CBY8 A0A1B0CMQ7 A0A2S2PDP3 A0A2S2P6J9 A0A2S2NWV3 X1X7V8 X1X2F5 J9LNE5 A0A1B0DHQ9 X1WQY4 J9LJG0 A0A0J7KTN8 A0A2S2NQE0 A0A2S2Q8X5 A0A224XT32 X1WK76 A0A2S2NXZ1 J9LNL0 X1X088 A0A1B0CRN6 A0A2S2R8S1 A0A2S2QG05 J9L3V2 A0A0J7MYU4 A0A2S2P9D3
EMBL
BABH01004047
BABH01004048
BABH01004049
BABH01004050
KQ460118
KPJ17710.1
+ More
NEVH01014359 PNF27827.1 NEVH01015822 PNF26627.1 AJWK01015895 LBMM01006314 KMQ90711.1 NWSH01002380 PCG68431.1 GGMS01016195 MBY85398.1 GEZM01016225 GEZM01016224 GEZM01016223 JAV91321.1 ABLF02024784 ABLF02024792 AJWK01010384 AJWK01005577 AJWK01005997 AJWK01019099 GGMR01014893 MBY27512.1 GGMR01012472 MBY25091.1 GGMR01009090 MBY21709.1 ABLF02017573 ABLF02054931 ABLF02024857 ABLF02059360 ABLF02001804 ABLF02035179 ABLF02062794 AJVK01061445 ABLF02042106 ABLF02016540 LBMM01003300 KMQ93691.1 GGMR01006776 MBY19395.1 GGMS01004984 MBY74187.1 GFTR01005305 JAW11121.1 ABLF02035261 ABLF02035909 ABLF02066026 GGMR01009313 MBY21932.1 ABLF02018955 ABLF02041521 AJWK01025019 GGMS01017182 MBY86385.1 GGMS01007463 MBY76666.1 ABLF02025592 LBMM01013507 KMQ85610.1 GGMR01013395 MBY26014.1
NEVH01014359 PNF27827.1 NEVH01015822 PNF26627.1 AJWK01015895 LBMM01006314 KMQ90711.1 NWSH01002380 PCG68431.1 GGMS01016195 MBY85398.1 GEZM01016225 GEZM01016224 GEZM01016223 JAV91321.1 ABLF02024784 ABLF02024792 AJWK01010384 AJWK01005577 AJWK01005997 AJWK01019099 GGMR01014893 MBY27512.1 GGMR01012472 MBY25091.1 GGMR01009090 MBY21709.1 ABLF02017573 ABLF02054931 ABLF02024857 ABLF02059360 ABLF02001804 ABLF02035179 ABLF02062794 AJVK01061445 ABLF02042106 ABLF02016540 LBMM01003300 KMQ93691.1 GGMR01006776 MBY19395.1 GGMS01004984 MBY74187.1 GFTR01005305 JAW11121.1 ABLF02035261 ABLF02035909 ABLF02066026 GGMR01009313 MBY21932.1 ABLF02018955 ABLF02041521 AJWK01025019 GGMS01017182 MBY86385.1 GGMS01007463 MBY76666.1 ABLF02025592 LBMM01013507 KMQ85610.1 GGMR01013395 MBY26014.1
Proteomes
PRIDE
Interpro
IPR006579
Pre_C2HC_dom
+ More
IPR036527 SCP2_sterol-bd_dom_sf
IPR039543 POX18/YhbT/NSL-TP1
IPR003033 SCP2_sterol-bd_dom
IPR019786 Zinc_finger_PHD-type_CS
IPR011011 Znf_FYVE_PHD
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR019787 Znf_PHD-finger
IPR000477 RT_dom
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR035979 RBD_domain_sf
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR036527 SCP2_sterol-bd_dom_sf
IPR039543 POX18/YhbT/NSL-TP1
IPR003033 SCP2_sterol-bd_dom
IPR019786 Zinc_finger_PHD-type_CS
IPR011011 Znf_FYVE_PHD
IPR001965 Znf_PHD
IPR013083 Znf_RING/FYVE/PHD
IPR019787 Znf_PHD-finger
IPR000477 RT_dom
IPR036691 Endo/exonu/phosph_ase_sf
IPR005135 Endo/exonuclease/phosphatase
IPR035979 RBD_domain_sf
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
SUPFAM
Gene 3D
ProteinModelPortal
H9JTE2
A0A194RK46
A0A2J7QGW8
A0A2J7QDG6
A0A1B0CK87
A0A0J7KKG5
+ More
A0A2A4J8Z7 A0A2S2R608 A0A1Y1N079 J9KE07 A0A1B0FV55 A0A1B0GHS3 A0A1B0CBY8 A0A1B0CMQ7 A0A2S2PDP3 A0A2S2P6J9 A0A2S2NWV3 X1X7V8 X1X2F5 J9LNE5 A0A1B0DHQ9 X1WQY4 J9LJG0 A0A0J7KTN8 A0A2S2NQE0 A0A2S2Q8X5 A0A224XT32 X1WK76 A0A2S2NXZ1 J9LNL0 X1X088 A0A1B0CRN6 A0A2S2R8S1 A0A2S2QG05 J9L3V2 A0A0J7MYU4 A0A2S2P9D3
A0A2A4J8Z7 A0A2S2R608 A0A1Y1N079 J9KE07 A0A1B0FV55 A0A1B0GHS3 A0A1B0CBY8 A0A1B0CMQ7 A0A2S2PDP3 A0A2S2P6J9 A0A2S2NWV3 X1X7V8 X1X2F5 J9LNE5 A0A1B0DHQ9 X1WQY4 J9LJG0 A0A0J7KTN8 A0A2S2NQE0 A0A2S2Q8X5 A0A224XT32 X1WK76 A0A2S2NXZ1 J9LNL0 X1X088 A0A1B0CRN6 A0A2S2R8S1 A0A2S2QG05 J9L3V2 A0A0J7MYU4 A0A2S2P9D3
PDB
5I1R
E-value=0.0134258,
Score=91
Ontologies
KEGG
PANTHER
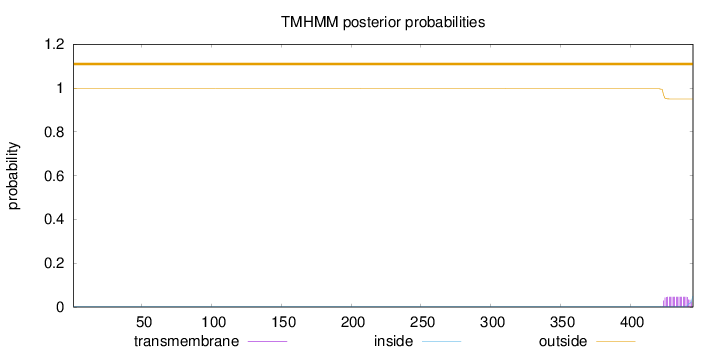
Topology
Length:
445
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.90163
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00364
outside
1 - 445
Population Genetic Test Statistics
Pi
165.782767
Theta
147.644333
Tajima's D
0.491814
CLR
0
CSRT
0.515274236288186
Interpretation
Uncertain
