Gene
KWMTBOMO13589 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA011000
Annotation
PREDICTED:_aminoacylase-1-like_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 2.48
Sequence
CDS
ATGCTCAGAATACATACAAGTTCACTCAAGAGTGACCCCAGCGTCTCGACCTTACAGAACTATCTTAGAATACGTTCAGTACATCCGAACGTTGACTACAATGAATGCATCAACTTCCTCAAAAATGAAGCTGAAAAGATTGGACTGCAAGTGCAAGTGGTCGAACCTTTGCCAAAGAAGCCGACGTTGGTAATGACCTGGTTAGGAGAGCAGCCTGAGCTGCCGAGCATATTACTCAACTCTCATATGGACGTGGTGCCTGTCTTCGAGAACAGCTGGAAGCATCCTCCCTTTGCTGCTGAAATAGAAGACAATGTAATTTACGCTAGAGGAGTTCAAGATATGAAGTCGGTTGGAATTCAGTACATTGAGGCCGTGAGGAGGCTGAAGGAAACCGGCGTCAAGCTCAAAAGGACTGTGCATCTGTCATTTGTACCCGACGAGGAGATTGGTGGCGATACTGGAATGGGGAAGTTCGTTCAGACGGACGTCTTCAAAAATATGAATGTCGGGTTCGCTCTCGATGAGGGCGTCGCATCCTCTAACGATGACTACCTGGTCTTCAATGGCGAAAGAATAATTTGGCATGTGAAAATCACTTGTCCCGGAAAATCCGGTCACGGTTCCCTTCTGCTCCCGGACAACTGCGGCGAAAAGTTGCGGTACATAATCGATAAGTTCATGGACCTGCGTCAGGAGTCCGTTAAAAAGTTGGCTGACAACCCACAGCTAACCATCGGTGACGTCACTAGCGTCAACCTCACCATGATCTCCGGAGGTATACAGAACAATGTGGTTCCTGAACAATTTACAGCCAACTTCGACCTTCGCATAGCTCTGAACGTCGATTTAAAAGAATTCGAAAATATGATTCAGAAATGGTGTACTGAAGCTGGTCGCGGCGTCACGTACGAGTTTAAGCAGAAGGATCCTTACACCACACCAACTCAGCTAGACGATGCCAACATTTACTGGAAAGCGTTCAAACAAACCGCACGGGAACTAGGAATGTCAATTAAGCCTCAAACGTTTACGGGAGGCACCGACAGCCGATACCTTCGGGAACTTGGCATCCCGGCGCTGGGCTTCTCGCCGATACACAACACCACGCCCGCGCTGCATGAGCACAACGAGCATCTAGGCTTGGACGTCTTCATCAATGGAATAGCGGTGTACCAGAGCATCATAAGGGCTGTAGCTAATGCCTAA
Protein
MLRIHTSSLKSDPSVSTLQNYLRIRSVHPNVDYNECINFLKNEAEKIGLQVQVVEPLPKKPTLVMTWLGEQPELPSILLNSHMDVVPVFENSWKHPPFAAEIEDNVIYARGVQDMKSVGIQYIEAVRRLKETGVKLKRTVHLSFVPDEEIGGDTGMGKFVQTDVFKNMNVGFALDEGVASSNDDYLVFNGERIIWHVKITCPGKSGHGSLLLPDNCGEKLRYIIDKFMDLRQESVKKLADNPQLTIGDVTSVNLTMISGGIQNNVVPEQFTANFDLRIALNVDLKEFENMIQKWCTEAGRGVTYEFKQKDPYTTPTQLDDANIYWKAFKQTARELGMSIKPQTFTGGTDSRYLRELGIPALGFSPIHNTTPALHEHNEHLGLDVFINGIAVYQSIIRAVANA
Summary
Uniprot
H9JN97
Q2F5U1
H9JN99
H9JN98
A0A2W1C250
A0A3S2TTR3
+ More
A0A194PSY0 A0A194PTR5 H9JLF1 A0A194QTV4 A0A212FLD6 A0A0P4VR91 A0A2H1VZW7 A0A139WBF2 W5JWJ1 A0A067R0Q5 T1DNA5 A0A2P8Y3J1 U5EYC3 A0A2M4BPI8 A0A2M4BQE3 A0A2J7RHR6 T1HYA1 W8ASK2 A0A154PAG0 A0A069DSV9 A0A0L7RA17 T1P9J4 A0A182SU41 B0WSY8 A0A2M3Z7X6 A0A084VVB7 V5G640 B4NF67 A0A182N000 A0A1Q3FN65 A0A087ZSU7 A0A310SSE3 A0A2M4AG56 B3P1M4 A0A182J7P4 A0A1W4VZI8 A0A182GM33 A0A182VVF1 A0A3B0K2X9 A0A182PU08 A0A1B0CBE6 A0A182I9W7 E9IDS2 A0A182X7H3 A0A182M7A4 Q16QR2 B4K4M9 A0A1L8E2H7 A0A0M4E350 A0A1L8E2D6 Q16QR1 B4LZT2 A0A182V259 B4HIS3 A0A182TFJ2 B4KH74 B4NJN5 A0A182XY72 B4G688 A0A182QZB9 D3TP85 Q7QEF5 B4JDK8 E2BIK0 A0A2A3EEL9 B3LV55 A0A1Y1NL69 A0A1A9UE48 B5DW61 A0A1A9X554 B4PKQ8 B4PN40 B4K1D6 A0A3B0K5C5 B3P7G8 A0A0M8ZT35 B5DTQ6 B3LVY1 A0A1B6FCZ8 B4R1C9 A0A1W4V321 A0A1B0A2X7 Q7JUX5 A0A1A9XKN7 Q9I7K3 B5DYA6 D3TPC1 A0A0L0C7Z4 Q9VCR2 A0A0L0C7Z3 A0A1A9UE50 A0A1W4UR39 B4HFA8 A0A224XM09
A0A194PSY0 A0A194PTR5 H9JLF1 A0A194QTV4 A0A212FLD6 A0A0P4VR91 A0A2H1VZW7 A0A139WBF2 W5JWJ1 A0A067R0Q5 T1DNA5 A0A2P8Y3J1 U5EYC3 A0A2M4BPI8 A0A2M4BQE3 A0A2J7RHR6 T1HYA1 W8ASK2 A0A154PAG0 A0A069DSV9 A0A0L7RA17 T1P9J4 A0A182SU41 B0WSY8 A0A2M3Z7X6 A0A084VVB7 V5G640 B4NF67 A0A182N000 A0A1Q3FN65 A0A087ZSU7 A0A310SSE3 A0A2M4AG56 B3P1M4 A0A182J7P4 A0A1W4VZI8 A0A182GM33 A0A182VVF1 A0A3B0K2X9 A0A182PU08 A0A1B0CBE6 A0A182I9W7 E9IDS2 A0A182X7H3 A0A182M7A4 Q16QR2 B4K4M9 A0A1L8E2H7 A0A0M4E350 A0A1L8E2D6 Q16QR1 B4LZT2 A0A182V259 B4HIS3 A0A182TFJ2 B4KH74 B4NJN5 A0A182XY72 B4G688 A0A182QZB9 D3TP85 Q7QEF5 B4JDK8 E2BIK0 A0A2A3EEL9 B3LV55 A0A1Y1NL69 A0A1A9UE48 B5DW61 A0A1A9X554 B4PKQ8 B4PN40 B4K1D6 A0A3B0K5C5 B3P7G8 A0A0M8ZT35 B5DTQ6 B3LVY1 A0A1B6FCZ8 B4R1C9 A0A1W4V321 A0A1B0A2X7 Q7JUX5 A0A1A9XKN7 Q9I7K3 B5DYA6 D3TPC1 A0A0L0C7Z4 Q9VCR2 A0A0L0C7Z3 A0A1A9UE50 A0A1W4UR39 B4HFA8 A0A224XM09
Pubmed
19121390
28756777
26354079
22118469
27129103
18362917
+ More
19820115 20920257 23761445 24845553 29403074 24495485 26334808 25315136 24438588 17994087 26483478 21282665 17510324 18057021 25244985 20353571 12364791 14747013 17210077 20798317 28004739 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605
19820115 20920257 23761445 24845553 29403074 24495485 26334808 25315136 24438588 17994087 26483478 21282665 17510324 18057021 25244985 20353571 12364791 14747013 17210077 20798317 28004739 15632085 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 26108605
EMBL
BABH01033304
DQ311332
ABD36276.1
BABH01033306
BABH01033307
BABH01033305
+ More
KZ149909 PZC78193.1 RSAL01000002 RVE55002.1 RVE55055.1 KQ459595 KPI95869.1 KQ459592 KPI96806.1 BABH01037200 BABH01037201 KQ461194 KPJ06971.1 AGBW02007778 OWR54568.1 GDKW01001674 JAI54921.1 ODYU01005458 SOQ46337.1 KQ971372 KYB25257.1 ADMH02000148 ETN67605.1 KK852801 KDR16327.1 GAMD01003060 JAA98530.1 PYGN01000970 PSN38838.1 GANO01002088 JAB57783.1 GGFJ01005848 MBW54989.1 GGFJ01005847 MBW54988.1 NEVH01003737 PNF40375.1 ACPB03023997 ACPB03025303 GAMC01014710 JAB91845.1 KQ434857 KZC08889.1 GBGD01001711 JAC87178.1 KQ414619 KOC67717.1 KA644835 AFP59464.1 DS232078 EDS34121.1 GGFM01003839 MBW24590.1 ATLV01017170 KE525157 KFB41911.1 GALX01002936 JAB65530.1 CH964251 EDW82934.1 GFDL01005996 JAV29049.1 KQ760974 OAD58563.1 GGFK01006396 MBW39717.1 CH954181 EDV49623.1 JXUM01073756 JXUM01073757 JXUM01073758 JXUM01073759 KQ562794 KXJ75120.1 OUUW01000013 SPP87673.1 AJWK01005087 APCN01001063 GL762535 EFZ21239.1 AXCM01002614 CH477736 EAT36742.1 CH933806 EDW15005.1 KRG01314.1 GFDF01001166 JAV12918.1 CP012523 ALC40178.1 GFDF01001181 JAV12903.1 EAT36741.1 CH940650 EDW68251.1 CH480815 EDW42720.1 CH933807 EDW13291.1 CH964272 EDW84997.1 CH479179 EDW23845.1 AXCN02001335 EZ423237 ADD19513.1 AAAB01008847 EAA06832.4 CH916368 EDW03378.1 GL448516 EFN84447.1 KZ288280 PBC29616.1 CH902617 EDV42527.1 GEZM01003849 JAV96527.1 CM000070 EDY67743.1 CM000160 EDW96817.1 EDW98095.1 CH917093 EDW04843.1 OUUW01000005 SPP81199.1 CH954182 EDV54057.1 KQ435903 KOX69073.1 CH476048 EDY71312.1 EDV41514.2 GECZ01021711 JAS48058.1 CM000364 EDX14015.1 AY119627 AAM50281.2 AE014297 AY047532 AAG22139.1 AAK77264.1 EDY67583.1 EZ423273 ADD19549.1 JRES01000781 KNC28371.1 AAF56094.2 AAN13926.1 AHN57477.1 KNC28366.1 EDW43286.1 GFTR01005568 JAW10858.1
KZ149909 PZC78193.1 RSAL01000002 RVE55002.1 RVE55055.1 KQ459595 KPI95869.1 KQ459592 KPI96806.1 BABH01037200 BABH01037201 KQ461194 KPJ06971.1 AGBW02007778 OWR54568.1 GDKW01001674 JAI54921.1 ODYU01005458 SOQ46337.1 KQ971372 KYB25257.1 ADMH02000148 ETN67605.1 KK852801 KDR16327.1 GAMD01003060 JAA98530.1 PYGN01000970 PSN38838.1 GANO01002088 JAB57783.1 GGFJ01005848 MBW54989.1 GGFJ01005847 MBW54988.1 NEVH01003737 PNF40375.1 ACPB03023997 ACPB03025303 GAMC01014710 JAB91845.1 KQ434857 KZC08889.1 GBGD01001711 JAC87178.1 KQ414619 KOC67717.1 KA644835 AFP59464.1 DS232078 EDS34121.1 GGFM01003839 MBW24590.1 ATLV01017170 KE525157 KFB41911.1 GALX01002936 JAB65530.1 CH964251 EDW82934.1 GFDL01005996 JAV29049.1 KQ760974 OAD58563.1 GGFK01006396 MBW39717.1 CH954181 EDV49623.1 JXUM01073756 JXUM01073757 JXUM01073758 JXUM01073759 KQ562794 KXJ75120.1 OUUW01000013 SPP87673.1 AJWK01005087 APCN01001063 GL762535 EFZ21239.1 AXCM01002614 CH477736 EAT36742.1 CH933806 EDW15005.1 KRG01314.1 GFDF01001166 JAV12918.1 CP012523 ALC40178.1 GFDF01001181 JAV12903.1 EAT36741.1 CH940650 EDW68251.1 CH480815 EDW42720.1 CH933807 EDW13291.1 CH964272 EDW84997.1 CH479179 EDW23845.1 AXCN02001335 EZ423237 ADD19513.1 AAAB01008847 EAA06832.4 CH916368 EDW03378.1 GL448516 EFN84447.1 KZ288280 PBC29616.1 CH902617 EDV42527.1 GEZM01003849 JAV96527.1 CM000070 EDY67743.1 CM000160 EDW96817.1 EDW98095.1 CH917093 EDW04843.1 OUUW01000005 SPP81199.1 CH954182 EDV54057.1 KQ435903 KOX69073.1 CH476048 EDY71312.1 EDV41514.2 GECZ01021711 JAS48058.1 CM000364 EDX14015.1 AY119627 AAM50281.2 AE014297 AY047532 AAG22139.1 AAK77264.1 EDY67583.1 EZ423273 ADD19549.1 JRES01000781 KNC28371.1 AAF56094.2 AAN13926.1 AHN57477.1 KNC28366.1 EDW43286.1 GFTR01005568 JAW10858.1
Proteomes
UP000005204
UP000283053
UP000053268
UP000053240
UP000007151
UP000007266
+ More
UP000000673 UP000027135 UP000245037 UP000235965 UP000015103 UP000076502 UP000053825 UP000095301 UP000075901 UP000002320 UP000030765 UP000007798 UP000075884 UP000005203 UP000008711 UP000075880 UP000192221 UP000069940 UP000249989 UP000075920 UP000268350 UP000075885 UP000092461 UP000075840 UP000076407 UP000075883 UP000008820 UP000009192 UP000092553 UP000008792 UP000075903 UP000001292 UP000075902 UP000076408 UP000008744 UP000075886 UP000007062 UP000001070 UP000008237 UP000242457 UP000007801 UP000078200 UP000001819 UP000091820 UP000002282 UP000053105 UP000000304 UP000092445 UP000092443 UP000000803 UP000037069
UP000000673 UP000027135 UP000245037 UP000235965 UP000015103 UP000076502 UP000053825 UP000095301 UP000075901 UP000002320 UP000030765 UP000007798 UP000075884 UP000005203 UP000008711 UP000075880 UP000192221 UP000069940 UP000249989 UP000075920 UP000268350 UP000075885 UP000092461 UP000075840 UP000076407 UP000075883 UP000008820 UP000009192 UP000092553 UP000008792 UP000075903 UP000001292 UP000075902 UP000076408 UP000008744 UP000075886 UP000007062 UP000001070 UP000008237 UP000242457 UP000007801 UP000078200 UP000001819 UP000091820 UP000002282 UP000053105 UP000000304 UP000092445 UP000092443 UP000000803 UP000037069
Interpro
ProteinModelPortal
H9JN97
Q2F5U1
H9JN99
H9JN98
A0A2W1C250
A0A3S2TTR3
+ More
A0A194PSY0 A0A194PTR5 H9JLF1 A0A194QTV4 A0A212FLD6 A0A0P4VR91 A0A2H1VZW7 A0A139WBF2 W5JWJ1 A0A067R0Q5 T1DNA5 A0A2P8Y3J1 U5EYC3 A0A2M4BPI8 A0A2M4BQE3 A0A2J7RHR6 T1HYA1 W8ASK2 A0A154PAG0 A0A069DSV9 A0A0L7RA17 T1P9J4 A0A182SU41 B0WSY8 A0A2M3Z7X6 A0A084VVB7 V5G640 B4NF67 A0A182N000 A0A1Q3FN65 A0A087ZSU7 A0A310SSE3 A0A2M4AG56 B3P1M4 A0A182J7P4 A0A1W4VZI8 A0A182GM33 A0A182VVF1 A0A3B0K2X9 A0A182PU08 A0A1B0CBE6 A0A182I9W7 E9IDS2 A0A182X7H3 A0A182M7A4 Q16QR2 B4K4M9 A0A1L8E2H7 A0A0M4E350 A0A1L8E2D6 Q16QR1 B4LZT2 A0A182V259 B4HIS3 A0A182TFJ2 B4KH74 B4NJN5 A0A182XY72 B4G688 A0A182QZB9 D3TP85 Q7QEF5 B4JDK8 E2BIK0 A0A2A3EEL9 B3LV55 A0A1Y1NL69 A0A1A9UE48 B5DW61 A0A1A9X554 B4PKQ8 B4PN40 B4K1D6 A0A3B0K5C5 B3P7G8 A0A0M8ZT35 B5DTQ6 B3LVY1 A0A1B6FCZ8 B4R1C9 A0A1W4V321 A0A1B0A2X7 Q7JUX5 A0A1A9XKN7 Q9I7K3 B5DYA6 D3TPC1 A0A0L0C7Z4 Q9VCR2 A0A0L0C7Z3 A0A1A9UE50 A0A1W4UR39 B4HFA8 A0A224XM09
A0A194PSY0 A0A194PTR5 H9JLF1 A0A194QTV4 A0A212FLD6 A0A0P4VR91 A0A2H1VZW7 A0A139WBF2 W5JWJ1 A0A067R0Q5 T1DNA5 A0A2P8Y3J1 U5EYC3 A0A2M4BPI8 A0A2M4BQE3 A0A2J7RHR6 T1HYA1 W8ASK2 A0A154PAG0 A0A069DSV9 A0A0L7RA17 T1P9J4 A0A182SU41 B0WSY8 A0A2M3Z7X6 A0A084VVB7 V5G640 B4NF67 A0A182N000 A0A1Q3FN65 A0A087ZSU7 A0A310SSE3 A0A2M4AG56 B3P1M4 A0A182J7P4 A0A1W4VZI8 A0A182GM33 A0A182VVF1 A0A3B0K2X9 A0A182PU08 A0A1B0CBE6 A0A182I9W7 E9IDS2 A0A182X7H3 A0A182M7A4 Q16QR2 B4K4M9 A0A1L8E2H7 A0A0M4E350 A0A1L8E2D6 Q16QR1 B4LZT2 A0A182V259 B4HIS3 A0A182TFJ2 B4KH74 B4NJN5 A0A182XY72 B4G688 A0A182QZB9 D3TP85 Q7QEF5 B4JDK8 E2BIK0 A0A2A3EEL9 B3LV55 A0A1Y1NL69 A0A1A9UE48 B5DW61 A0A1A9X554 B4PKQ8 B4PN40 B4K1D6 A0A3B0K5C5 B3P7G8 A0A0M8ZT35 B5DTQ6 B3LVY1 A0A1B6FCZ8 B4R1C9 A0A1W4V321 A0A1B0A2X7 Q7JUX5 A0A1A9XKN7 Q9I7K3 B5DYA6 D3TPC1 A0A0L0C7Z4 Q9VCR2 A0A0L0C7Z3 A0A1A9UE50 A0A1W4UR39 B4HFA8 A0A224XM09
PDB
1Q7L
E-value=9.52135e-51,
Score=506
Ontologies
PATHWAY
GO
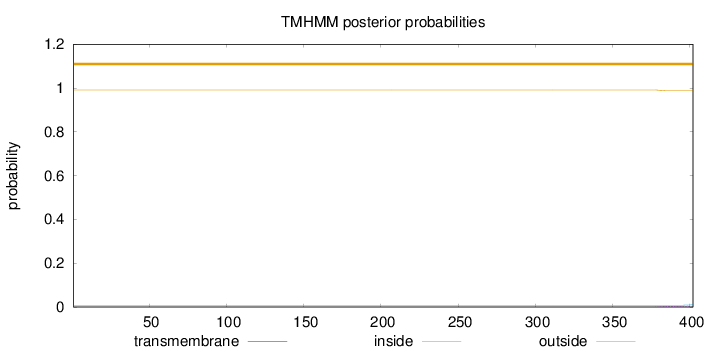
Topology
Length:
402
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.05547
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00846
outside
1 - 402
Population Genetic Test Statistics
Pi
261.815351
Theta
156.578276
Tajima's D
2.189749
CLR
0.329471
CSRT
0.911954402279886
Interpretation
Uncertain
