Gene
KWMTBOMO13584
Annotation
PREDICTED:_membrane-bound_transcription_factor_site-2_protease_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.878
Sequence
CDS
ATGCACTATCCATATTATGCATTTTTAGAAGGAACGGGCTTAAAATTTGGAATACTGAACTTTTCTTGGACGACAACTGCTTTTAACAGATGCCTTTATAGATGGAGTAAGAATTTGTCTCGAATACTCAGAAAATGGTTCACGTGCGGTTACATTCTTGTAATCTGGATATTCTTGCCTTTCGCTATATGGACTTTGCTCTCATTTATTTTTGAGCATTTTTATGATACAATACAGCTCAATAATGTACCAGAAGTCAAAGCAATGCTCCCTGGTATAAATATCCCGACATCAGACTTTTGGATTTATTTTCTAGCTATAGCATTGAGCACTATATTCCATGAACTAGGACATGCTCTTGCAGCTGCTCAAGAGGATGTTCAAATAATAGCAGTAGGAGTTTATGTATTTACTATTATACCAGTTGCATTTGTAGACTTGAATACTGATCAATTAAAGAATCTTGCTGTCACCAAAAGATTGAGGATTTACTGTGCTGGAGTGTGGCATAATATTGCAATTGCATTTATGGCTATCTTATTATTTTTCTCTGCTCCGATACTGTTTAATATTGCATATGAAACGGGAACGGGTGTAAGAGTCACTGACCTTACAGAAGACTCACCATTAAAGGACAGCATAGGCTTGCACAAAGATGATGTAATAACATCAATAAACAACTGTAAGATCAAAGTTGCAAGTGATTGGATGCAATGTTTGCAATTAGCACATCAGAGGTATGGAATTTGTACAAGTGCTGAATTTGTTGCCCAAAATGATGAGATCATGATGGAGAACATAAAAGAGAATGATGTGGTAGAATGTTGTAGACCAGAAGAAGCATATAATCATTGTTTTGAGTATATGGAGCCTAAAGTAGCATCTGATTCAGTTTTACCCGGACAATACTCATGCCTCAAACCCAGAGATATGGTGAAGAATCAGTTTGTAAAATGTACAGAATCTGGAGGATACTCTTGCCCAAGAGCCATGCATTGTATGAGACCTTCACTAAATAATGTATCCTACCTCCTAATAATTGAACGAAAGGAGAACAATCCAGTTCTGTATTTAGGGTTGCCGTATGATTTACACAAAACTGTATTTGTCGACCAGTATTTCCCCAGACTGAAAGTATTTTCATTTTTTTCCCCTTCTCGGTTTGAGAAGCTCCTTAGATACATATTCATATTTTCTATGGGTATCGCATTTTTGAACGTCTTACCATGTTACAGTATGGATGGACACCATATAGCTCGTAACTTGATACAAATTTTAGCTAAATATTTAAATAAAAATGGTGATTTTGTAACATTCTTTACAGTATTCACTGTAGTTATAGGCACAGGTGTGACTGGGCCCATTTTAATATATTTGTTCTACAAAGCTGTATATGAAGATAATTAG
Protein
MHYPYYAFLEGTGLKFGILNFSWTTTAFNRCLYRWSKNLSRILRKWFTCGYILVIWIFLPFAIWTLLSFIFEHFYDTIQLNNVPEVKAMLPGINIPTSDFWIYFLAIALSTIFHELGHALAAAQEDVQIIAVGVYVFTIIPVAFVDLNTDQLKNLAVTKRLRIYCAGVWHNIAIAFMAILLFFSAPILFNIAYETGTGVRVTDLTEDSPLKDSIGLHKDDVITSINNCKIKVASDWMQCLQLAHQRYGICTSAEFVAQNDEIMMENIKENDVVECCRPEEAYNHCFEYMEPKVASDSVLPGQYSCLKPRDMVKNQFVKCTESGGYSCPRAMHCMRPSLNNVSYLLIIERKENNPVLYLGLPYDLHKTVFVDQYFPRLKVFSFFSPSRFEKLLRYIFIFSMGIAFLNVLPCYSMDGHHIARNLIQILAKYLNKNGDFVTFFTVFTVVIGTGVTGPILIYLFYKAVYEDN
Summary
Uniprot
A0A2A4J2G8
A0A2W1BSZ7
A0A2H1X462
A0A3S2NAK5
A0A194PSY5
A0A0L7KSP4
+ More
A0A194REV6 A0A182M2E2 A0A182RYZ0 A0A182J270 A0A182YL90 A0A182K3Z6 A0A182SZ79 A0A182QL32 A0A084WJ88 A0A182W1W3 W5JF86 A0A182FUM1 A0A182P6Y6 A0A182NSY7 A0A1W4WTA3 B0WTG2 D6WA43 A0A182TQM5 Q7PZA9 A0A182HHC9 A0A182XGR2 A0A182V141 Q16J11 Q16MF4 A0A1S4FVS7 A0A087ZWY1 A0A2A3ESB1 A0A026W0X0 E2BFC3 A0A3L8DMD5 E1ZWR7 A0A0J7K186 A0A195FP35 A0A195BDU0 E9I8D3 A0A151XDU3 A0A1B6EDP2 A0A0A9XDY4 A0A310STY4 A0A158NIL9 F4WS92 A0A195CKF4 A0A151J1Z6 A0A0N1ITI3 A0A0L7RGF7 A0A2S2R0R5 A0A067RFH8 A0A2J7PEA6 A0A1L8DTQ4 A0A1B6LD29 A0A224XPU2 A0A1Y1KAX4 A0A1B6HK06 A0A069DUI3 A0A2H8TEB5 A0A1B6F1T7 A0A023F1E0 A0A0V0GAT6 J9JSR0 A0A0A1WTT7 A0A2S2NSN7 A0A293LH98 A0A0K8UY76 T1PJ03 A0A1I8NG20 A0A0M4EI54 W8BV72 A0A1B0C675 T1J3H3 K1PKA9 B3MCV2 A0A336M4L4 W4XCS3 A0A1A9XTY2 A0A1I8Q6A4 A0A1W4V1A3 A0A3B0J5M1 A0A1B6E0Q3 B4KUF6 A0A0C9RAI9 Q28XC1 B4H5A4 B4N5P9 A0A1B0A9S9 A0A1S3JB99 A0A1S3JA13 B4P606 A0A1B0GBB0 A0A1A9WW47 B4QBP9 Q7JZ56 A0A1J1IWJ0
A0A194REV6 A0A182M2E2 A0A182RYZ0 A0A182J270 A0A182YL90 A0A182K3Z6 A0A182SZ79 A0A182QL32 A0A084WJ88 A0A182W1W3 W5JF86 A0A182FUM1 A0A182P6Y6 A0A182NSY7 A0A1W4WTA3 B0WTG2 D6WA43 A0A182TQM5 Q7PZA9 A0A182HHC9 A0A182XGR2 A0A182V141 Q16J11 Q16MF4 A0A1S4FVS7 A0A087ZWY1 A0A2A3ESB1 A0A026W0X0 E2BFC3 A0A3L8DMD5 E1ZWR7 A0A0J7K186 A0A195FP35 A0A195BDU0 E9I8D3 A0A151XDU3 A0A1B6EDP2 A0A0A9XDY4 A0A310STY4 A0A158NIL9 F4WS92 A0A195CKF4 A0A151J1Z6 A0A0N1ITI3 A0A0L7RGF7 A0A2S2R0R5 A0A067RFH8 A0A2J7PEA6 A0A1L8DTQ4 A0A1B6LD29 A0A224XPU2 A0A1Y1KAX4 A0A1B6HK06 A0A069DUI3 A0A2H8TEB5 A0A1B6F1T7 A0A023F1E0 A0A0V0GAT6 J9JSR0 A0A0A1WTT7 A0A2S2NSN7 A0A293LH98 A0A0K8UY76 T1PJ03 A0A1I8NG20 A0A0M4EI54 W8BV72 A0A1B0C675 T1J3H3 K1PKA9 B3MCV2 A0A336M4L4 W4XCS3 A0A1A9XTY2 A0A1I8Q6A4 A0A1W4V1A3 A0A3B0J5M1 A0A1B6E0Q3 B4KUF6 A0A0C9RAI9 Q28XC1 B4H5A4 B4N5P9 A0A1B0A9S9 A0A1S3JB99 A0A1S3JA13 B4P606 A0A1B0GBB0 A0A1A9WW47 B4QBP9 Q7JZ56 A0A1J1IWJ0
Pubmed
28756777
26354079
26227816
25244985
24438588
20920257
+ More
23761445 18362917 19820115 12364791 14747013 17210077 17510324 24508170 20798317 30249741 21282665 25401762 26823975 21347285 21719571 24845553 28004739 26334808 25474469 25830018 25315136 24495485 22992520 17994087 15632085 17550304 22936249 10731132 11832248 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
23761445 18362917 19820115 12364791 14747013 17210077 17510324 24508170 20798317 30249741 21282665 25401762 26823975 21347285 21719571 24845553 28004739 26334808 25474469 25830018 25315136 24495485 22992520 17994087 15632085 17550304 22936249 10731132 11832248 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356
EMBL
NWSH01003415
PCG66347.1
KZ149909
PZC78188.1
ODYU01012883
SOQ59414.1
+ More
RSAL01000146 RSAL01000102 RVE45916.1 RVE47439.1 KQ459595 KPI95874.1 JTDY01006347 KOB66086.1 KQ460313 KPJ16112.1 AXCM01003078 AXCN02002132 ATLV01023981 KE525347 KFB50282.1 ADMH02001316 ETN63032.1 DS232085 EDS34400.1 KQ971312 EEZ98063.1 AAAB01008986 EAA00371.4 APCN01002406 CH478039 EAT34258.1 CH477863 EAT35505.1 KZ288189 PBC34618.1 KK107503 EZA49658.1 GL448009 EFN85583.1 QOIP01000006 RLU21476.1 GL434901 EFN74356.1 LBMM01017826 KMQ83956.1 KQ981430 KYN41704.1 KQ976513 KYM82372.1 GL761538 EFZ23169.1 KQ982254 KYQ58541.1 GEDC01015788 GEDC01001240 JAS21510.1 JAS36058.1 GBHO01026596 GBHO01026595 GBRD01017687 GDHC01001066 JAG17008.1 JAG17009.1 JAG48140.1 JAQ17563.1 KQ759888 OAD62118.1 ADTU01016949 GL888307 EGI62928.1 KQ977634 KYN01210.1 KQ980495 KYN15803.1 KQ435794 KOX73944.1 KQ414597 KOC69888.1 GGMS01014372 MBY83575.1 KK852498 KDR22611.1 NEVH01026106 PNF14659.1 GFDF01004248 JAV09836.1 GEBQ01018396 JAT21581.1 GFTR01006253 JAW10173.1 GEZM01087519 JAV58652.1 GECU01032811 JAS74895.1 GBGD01001274 JAC87615.1 GFXV01000247 MBW12052.1 GECZ01025635 JAS44134.1 GBBI01003903 JAC14809.1 GECL01001751 JAP04373.1 ABLF02034746 GBXI01012499 JAD01793.1 GGMR01007528 MBY20147.1 GFWV01002518 MAA27248.1 GDHF01021044 JAI31270.1 KA648659 AFP63288.1 CP012524 ALC40705.1 GAMC01009349 JAB97206.1 JXJN01026454 JH431826 JH817204 EKC19329.1 CH902619 EDV37354.1 UFQT01000552 SSX25206.1 AAGJ04118672 OUUW01000001 SPP74972.1 GEDC01022094 GEDC01005809 JAS15204.1 JAS31489.1 CH933808 EDW10743.1 GBYB01003831 JAG73598.1 CM000071 EAL26395.1 CH479210 EDW32940.1 CH964154 EDW79688.1 CM000158 EDW90881.1 CCAG010011567 CM000362 CM002911 EDX06666.1 KMY93042.1 AE013599 AY071529 AF441757 AAF58617.2 AAL49151.1 AAM20921.1 CVRI01000063 CRL04581.1
RSAL01000146 RSAL01000102 RVE45916.1 RVE47439.1 KQ459595 KPI95874.1 JTDY01006347 KOB66086.1 KQ460313 KPJ16112.1 AXCM01003078 AXCN02002132 ATLV01023981 KE525347 KFB50282.1 ADMH02001316 ETN63032.1 DS232085 EDS34400.1 KQ971312 EEZ98063.1 AAAB01008986 EAA00371.4 APCN01002406 CH478039 EAT34258.1 CH477863 EAT35505.1 KZ288189 PBC34618.1 KK107503 EZA49658.1 GL448009 EFN85583.1 QOIP01000006 RLU21476.1 GL434901 EFN74356.1 LBMM01017826 KMQ83956.1 KQ981430 KYN41704.1 KQ976513 KYM82372.1 GL761538 EFZ23169.1 KQ982254 KYQ58541.1 GEDC01015788 GEDC01001240 JAS21510.1 JAS36058.1 GBHO01026596 GBHO01026595 GBRD01017687 GDHC01001066 JAG17008.1 JAG17009.1 JAG48140.1 JAQ17563.1 KQ759888 OAD62118.1 ADTU01016949 GL888307 EGI62928.1 KQ977634 KYN01210.1 KQ980495 KYN15803.1 KQ435794 KOX73944.1 KQ414597 KOC69888.1 GGMS01014372 MBY83575.1 KK852498 KDR22611.1 NEVH01026106 PNF14659.1 GFDF01004248 JAV09836.1 GEBQ01018396 JAT21581.1 GFTR01006253 JAW10173.1 GEZM01087519 JAV58652.1 GECU01032811 JAS74895.1 GBGD01001274 JAC87615.1 GFXV01000247 MBW12052.1 GECZ01025635 JAS44134.1 GBBI01003903 JAC14809.1 GECL01001751 JAP04373.1 ABLF02034746 GBXI01012499 JAD01793.1 GGMR01007528 MBY20147.1 GFWV01002518 MAA27248.1 GDHF01021044 JAI31270.1 KA648659 AFP63288.1 CP012524 ALC40705.1 GAMC01009349 JAB97206.1 JXJN01026454 JH431826 JH817204 EKC19329.1 CH902619 EDV37354.1 UFQT01000552 SSX25206.1 AAGJ04118672 OUUW01000001 SPP74972.1 GEDC01022094 GEDC01005809 JAS15204.1 JAS31489.1 CH933808 EDW10743.1 GBYB01003831 JAG73598.1 CM000071 EAL26395.1 CH479210 EDW32940.1 CH964154 EDW79688.1 CM000158 EDW90881.1 CCAG010011567 CM000362 CM002911 EDX06666.1 KMY93042.1 AE013599 AY071529 AF441757 AAF58617.2 AAL49151.1 AAM20921.1 CVRI01000063 CRL04581.1
Proteomes
UP000218220
UP000283053
UP000053268
UP000037510
UP000053240
UP000075883
+ More
UP000075900 UP000075880 UP000076408 UP000075881 UP000075901 UP000075886 UP000030765 UP000075920 UP000000673 UP000069272 UP000075885 UP000075884 UP000192223 UP000002320 UP000007266 UP000075902 UP000007062 UP000075840 UP000076407 UP000075903 UP000008820 UP000005203 UP000242457 UP000053097 UP000008237 UP000279307 UP000000311 UP000036403 UP000078541 UP000078540 UP000075809 UP000005205 UP000007755 UP000078542 UP000078492 UP000053105 UP000053825 UP000027135 UP000235965 UP000007819 UP000095301 UP000092553 UP000092460 UP000005408 UP000007801 UP000007110 UP000092443 UP000095300 UP000192221 UP000268350 UP000009192 UP000001819 UP000008744 UP000007798 UP000092445 UP000085678 UP000002282 UP000092444 UP000091820 UP000000304 UP000000803 UP000183832
UP000075900 UP000075880 UP000076408 UP000075881 UP000075901 UP000075886 UP000030765 UP000075920 UP000000673 UP000069272 UP000075885 UP000075884 UP000192223 UP000002320 UP000007266 UP000075902 UP000007062 UP000075840 UP000076407 UP000075903 UP000008820 UP000005203 UP000242457 UP000053097 UP000008237 UP000279307 UP000000311 UP000036403 UP000078541 UP000078540 UP000075809 UP000005205 UP000007755 UP000078542 UP000078492 UP000053105 UP000053825 UP000027135 UP000235965 UP000007819 UP000095301 UP000092553 UP000092460 UP000005408 UP000007801 UP000007110 UP000092443 UP000095300 UP000192221 UP000268350 UP000009192 UP000001819 UP000008744 UP000007798 UP000092445 UP000085678 UP000002282 UP000092444 UP000091820 UP000000304 UP000000803 UP000183832
PRIDE
Pfam
PF02163 Peptidase_M50
SUPFAM
SSF50156
SSF50156
ProteinModelPortal
A0A2A4J2G8
A0A2W1BSZ7
A0A2H1X462
A0A3S2NAK5
A0A194PSY5
A0A0L7KSP4
+ More
A0A194REV6 A0A182M2E2 A0A182RYZ0 A0A182J270 A0A182YL90 A0A182K3Z6 A0A182SZ79 A0A182QL32 A0A084WJ88 A0A182W1W3 W5JF86 A0A182FUM1 A0A182P6Y6 A0A182NSY7 A0A1W4WTA3 B0WTG2 D6WA43 A0A182TQM5 Q7PZA9 A0A182HHC9 A0A182XGR2 A0A182V141 Q16J11 Q16MF4 A0A1S4FVS7 A0A087ZWY1 A0A2A3ESB1 A0A026W0X0 E2BFC3 A0A3L8DMD5 E1ZWR7 A0A0J7K186 A0A195FP35 A0A195BDU0 E9I8D3 A0A151XDU3 A0A1B6EDP2 A0A0A9XDY4 A0A310STY4 A0A158NIL9 F4WS92 A0A195CKF4 A0A151J1Z6 A0A0N1ITI3 A0A0L7RGF7 A0A2S2R0R5 A0A067RFH8 A0A2J7PEA6 A0A1L8DTQ4 A0A1B6LD29 A0A224XPU2 A0A1Y1KAX4 A0A1B6HK06 A0A069DUI3 A0A2H8TEB5 A0A1B6F1T7 A0A023F1E0 A0A0V0GAT6 J9JSR0 A0A0A1WTT7 A0A2S2NSN7 A0A293LH98 A0A0K8UY76 T1PJ03 A0A1I8NG20 A0A0M4EI54 W8BV72 A0A1B0C675 T1J3H3 K1PKA9 B3MCV2 A0A336M4L4 W4XCS3 A0A1A9XTY2 A0A1I8Q6A4 A0A1W4V1A3 A0A3B0J5M1 A0A1B6E0Q3 B4KUF6 A0A0C9RAI9 Q28XC1 B4H5A4 B4N5P9 A0A1B0A9S9 A0A1S3JB99 A0A1S3JA13 B4P606 A0A1B0GBB0 A0A1A9WW47 B4QBP9 Q7JZ56 A0A1J1IWJ0
A0A194REV6 A0A182M2E2 A0A182RYZ0 A0A182J270 A0A182YL90 A0A182K3Z6 A0A182SZ79 A0A182QL32 A0A084WJ88 A0A182W1W3 W5JF86 A0A182FUM1 A0A182P6Y6 A0A182NSY7 A0A1W4WTA3 B0WTG2 D6WA43 A0A182TQM5 Q7PZA9 A0A182HHC9 A0A182XGR2 A0A182V141 Q16J11 Q16MF4 A0A1S4FVS7 A0A087ZWY1 A0A2A3ESB1 A0A026W0X0 E2BFC3 A0A3L8DMD5 E1ZWR7 A0A0J7K186 A0A195FP35 A0A195BDU0 E9I8D3 A0A151XDU3 A0A1B6EDP2 A0A0A9XDY4 A0A310STY4 A0A158NIL9 F4WS92 A0A195CKF4 A0A151J1Z6 A0A0N1ITI3 A0A0L7RGF7 A0A2S2R0R5 A0A067RFH8 A0A2J7PEA6 A0A1L8DTQ4 A0A1B6LD29 A0A224XPU2 A0A1Y1KAX4 A0A1B6HK06 A0A069DUI3 A0A2H8TEB5 A0A1B6F1T7 A0A023F1E0 A0A0V0GAT6 J9JSR0 A0A0A1WTT7 A0A2S2NSN7 A0A293LH98 A0A0K8UY76 T1PJ03 A0A1I8NG20 A0A0M4EI54 W8BV72 A0A1B0C675 T1J3H3 K1PKA9 B3MCV2 A0A336M4L4 W4XCS3 A0A1A9XTY2 A0A1I8Q6A4 A0A1W4V1A3 A0A3B0J5M1 A0A1B6E0Q3 B4KUF6 A0A0C9RAI9 Q28XC1 B4H5A4 B4N5P9 A0A1B0A9S9 A0A1S3JB99 A0A1S3JA13 B4P606 A0A1B0GBB0 A0A1A9WW47 B4QBP9 Q7JZ56 A0A1J1IWJ0
Ontologies
GO
PANTHER
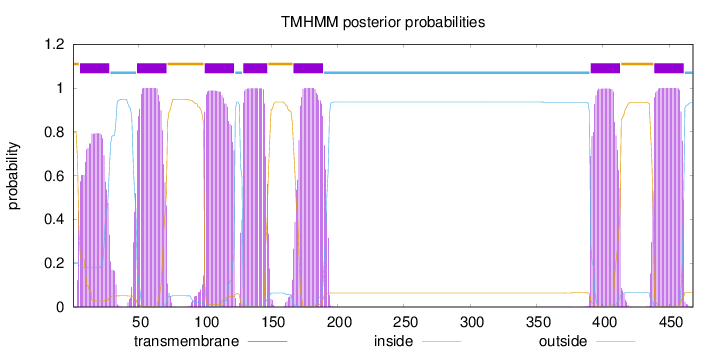
Topology
Length:
468
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
147.7269
Exp number, first 60 AAs:
30.07804
Total prob of N-in:
0.20355
POSSIBLE N-term signal
sequence
outside
1 - 5
TMhelix
6 - 28
inside
29 - 48
TMhelix
49 - 71
outside
72 - 99
TMhelix
100 - 122
inside
123 - 128
TMhelix
129 - 147
outside
148 - 166
TMhelix
167 - 189
inside
190 - 390
TMhelix
391 - 413
outside
414 - 438
TMhelix
439 - 461
inside
462 - 468
Population Genetic Test Statistics
Pi
232.815026
Theta
209.130534
Tajima's D
0.356541
CLR
0.167024
CSRT
0.466426678666067
Interpretation
Uncertain
