Gene
KWMTBOMO13578
Pre Gene Modal
BGIBMGA010965
Annotation
PREDICTED:_exostosin-2_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 3.305
Sequence
CDS
ATGGCTGCTGACTTGCTGTTGAAAGCGAGCAAGCATTCTAAGTATTTACCGTATAGAAATTTTTTGTATCATAGATTTTTTGTGGGACTGCTCTTGTTTATAGTGATATCTTTAGTGTTAACAGTGGTGTTCAATGCGTTTAGTGGTCCTTTGCCGCGGACTAGTGTTTACGAGTCTATTGATTTGAATAAAAGAAGCAAAGTGGTATTATCACGCAATGCTCCGTTTTCGCAGATTCGATCTGCGAATTGTACATATTGGGATTGCTTTGATGTCTACAAATGCGGGAGAGGAGGTCATGATAAGATAACCATATACATTTATCCGTTAAAAGATTATCAGACTGAGCATGGGAATTCAATATCGCAATTTTCTAGAGAATTTTACTTGATATTAGATACAATAAAACACAGCAAGTACTATACGACAAACCCAGAGGAAGCGTGTTTGCTGGTCCCTAGCATAGACACATTAAATCAAGCAAGTTTCTCTAAGAGTGAAGTGTCACAAGCTTTGCAATCTCTATCTCATTGGAATAATGGTGAAAACCACCTACTGTTCAACATGCTGCCTGGATCACCGCCTAATTACAATCCAGTCATCGACCTCAATACTTCAAAAGCAATTATAGCAGGTGCTGGATTTAATACATGGACTTTCCGCTATGGCTTTGACATTTCTGTACCAGTGTATAGCCAAATTGCTAAAAATATTGATAATACACAACCAAAAAAGAAAAACTATCTGATAATATCAGCGCAACTTAACATCCCCGAGAATTATCTTGAAGATTTACAGAGTACTGCATCGTCGTCTAATGATTTACTATTACTCGACGAATGTAAAACTGGGGATGTACTTGATTACACTAAAAGATGTGAATACACCACAGGCCGTGAGTTTAATTACCCAGATATATTGAAAGAGGGCTTGTTTTGTTTGGTTGTAAGAAGTGCAAGACTTACTCATTCATTATTAATGGATGTTATTGCCTCACAGTGCATACCAGTGGTTATATCGGATTCTGTTGTTATGCCCTTCAATTCTCATTTAGACTGGCATAGAATAGCAATGTTCATACCAGAAGATAACATAAAGAATTTGTTAAAGATCATCAGTTCAGTCAGTAATGAGAGACGTGGTGAATTGTTTTGGCAATTGCGTTGGGTTTATGATAAATATTTTGCGAGTATAGAGAAGATAACATTAGCAACATTAGAAATAATTAACGAGAAAGTGTTCCCGTTGGCAGCTAGGACATATGAAGAATGGAACATGCCAGAACATTTGTATGGTCCATCGAATCCACTATTCCTACCAGTGACAGCGCCGAAAGCACCAGGCTTTACTGCGGTCATATTAACGTACGACCGGGTCGAAAGTCTGTTTACGCTGGTCAAGAAACTAGTCCGCACCCCGAGCTTAGCCAAGATCCTTGTCATATGGAACAACCAAAAAAAAGCGCCTCCACCATCCGCCCAGTGGCCGGTCATCAATAAACCGTTAAAAGTGATACGGACGAAAGAAAACAAATTGTCGAACAGATTCTTTCCATACGACGAGATCGAAACGGAATGTCAGTTGACTATCGACGATGACATTGTCATGTTGACGCCCGACGAACTGGAGTTCGGTTTCGACGTATGGCGTGAGTTTCCGGACCGGATCGTCGGGTTCCCTTCGAGGCTCCATCTCTGGGATAACGCGACAAATTCGTGGCGTTACCACAGCGAATGGACTAATCAGATTTCTATGGTTTTAACCGGAGCCGCGTTCCATCACAAGATCTGGTCCTGGTACTACACGTATAAGATGCCGTCCGAGATCAGAACGTGGGTGGACGACAACTTCAATTGCGAAGACATAGCCATGAACTTCCTCGTCGCTAACGTGACCAGAAAACCTCCAATCAAGGTTACACCCCGTAAGAAGTTCAAGTGTCCGGAGTGCACCAACACGGAGATGCTGTCAGCGGACGCGCAGCACATGGCGCAGCGCTCGGCCTGCGTGGGGCGCTTCGCCTCCGCCTACGGGCACATGGCGCTCCGCCCCGTGGAGTTCCGCGCCGACCCGCTGCAGTACCGGGAGGCGGGGGCCGGCGTGCCGCACGCCTACCCCGACATCGGCGCCCTCTAG
Protein
MAADLLLKASKHSKYLPYRNFLYHRFFVGLLLFIVISLVLTVVFNAFSGPLPRTSVYESIDLNKRSKVVLSRNAPFSQIRSANCTYWDCFDVYKCGRGGHDKITIYIYPLKDYQTEHGNSISQFSREFYLILDTIKHSKYYTTNPEEACLLVPSIDTLNQASFSKSEVSQALQSLSHWNNGENHLLFNMLPGSPPNYNPVIDLNTSKAIIAGAGFNTWTFRYGFDISVPVYSQIAKNIDNTQPKKKNYLIISAQLNIPENYLEDLQSTASSSNDLLLLDECKTGDVLDYTKRCEYTTGREFNYPDILKEGLFCLVVRSARLTHSLLMDVIASQCIPVVISDSVVMPFNSHLDWHRIAMFIPEDNIKNLLKIISSVSNERRGELFWQLRWVYDKYFASIEKITLATLEIINEKVFPLAARTYEEWNMPEHLYGPSNPLFLPVTAPKAPGFTAVILTYDRVESLFTLVKKLVRTPSLAKILVIWNNQKKAPPPSAQWPVINKPLKVIRTKENKLSNRFFPYDEIETECQLTIDDDIVMLTPDELEFGFDVWREFPDRIVGFPSRLHLWDNATNSWRYHSEWTNQISMVLTGAAFHHKIWSWYYTYKMPSEIRTWVDDNFNCEDIAMNFLVANVTRKPPIKVTPRKKFKCPECTNTEMLSADAQHMAQRSACVGRFASAYGHMALRPVEFRADPLQYREAGAGVPHAYPDIGAL
Summary
Similarity
Belongs to the glycosyltransferase 47 family.
Uniprot
A0A2A4JMS1
A0A3S2LG08
A0A194RE51
A0A194PSD4
H9JN62
A0A2W1BX13
+ More
A0A212EWC9 A0A2H1WLY5 A0A2J7PQE6 A0A067REC1 A0A1Y1MFF0 A0A1Y1MF67 A0A146KMH8 A0A023F2Q0 A0A224XKR0 A0A0P4W4K8 A0A0V0GAA6 A0A069DVV8 Q0IFF4 A0A1B6D987 A0A1S4FBE3 A0A182GWM5 E0VWK8 A0A026X0R1 D6WCJ9 A0A182J5K6 A0A0J7NYK0 A0A336L2W6 A0A182R6F5 A0A182W3Y6 A0A182PFE8 A0A182T8P1 A0A158P279 A0A182MRP2 E9IV14 A0A151X210 A0A195FJA5 A0A182KJZ3 A0A182FJC8 W5JLS5 A0A182UYB6 U5EZH6 Q7PS14 A0A182XPP4 A0A0C9QQG9 A0A182HIM8 A0A0M9A227 A0A2M4BG34 A0A087ZZZ9 A0A154P3R5 U4UA52 A0A182YCN9 A0A1Q3FHA4 R7UWJ4 A0A1J1ITR8 A0A3P8UQE7 E2B382 L5KY03 W8CAA2 A0A3P9HQH6 K9J2H1 A0A3B0IYF4 A0A1S3KGP8 A0A3B3HCC2 A0A3Q4HKX7 A0A3B3CMJ3 I3JFZ6 A0A3P9KT00 A0A3P8RDI6 H3BD84 A0A2U9BCK2 V9K9N8 G1NDI8 A0A3B4FH49 A0A3Q2VLB3 A0A3Q3NAA6 B4LNV8 A0A3Q3N858 Q291L5 H2V525 A0A3B5AVG4 A0A0P7XFA4 A0A3S2UMX4 A0A3P9AT73 A0A2D0RV59 A0A1W5A662 A0A1A8RRU8 A0A1A8QD73 A0A3Q7R1J6 A0A3Q1EX73 A0A3Q7PVY4 A0A0F8AZG5 B4MRE0 A0A1V4KED0 A0A1A8EBW2 A0A1A8KLQ0 A0A3P8XZT3 A0A1A8BD97 A0A1A8V5M8 A0A3P8TCH4
A0A212EWC9 A0A2H1WLY5 A0A2J7PQE6 A0A067REC1 A0A1Y1MFF0 A0A1Y1MF67 A0A146KMH8 A0A023F2Q0 A0A224XKR0 A0A0P4W4K8 A0A0V0GAA6 A0A069DVV8 Q0IFF4 A0A1B6D987 A0A1S4FBE3 A0A182GWM5 E0VWK8 A0A026X0R1 D6WCJ9 A0A182J5K6 A0A0J7NYK0 A0A336L2W6 A0A182R6F5 A0A182W3Y6 A0A182PFE8 A0A182T8P1 A0A158P279 A0A182MRP2 E9IV14 A0A151X210 A0A195FJA5 A0A182KJZ3 A0A182FJC8 W5JLS5 A0A182UYB6 U5EZH6 Q7PS14 A0A182XPP4 A0A0C9QQG9 A0A182HIM8 A0A0M9A227 A0A2M4BG34 A0A087ZZZ9 A0A154P3R5 U4UA52 A0A182YCN9 A0A1Q3FHA4 R7UWJ4 A0A1J1ITR8 A0A3P8UQE7 E2B382 L5KY03 W8CAA2 A0A3P9HQH6 K9J2H1 A0A3B0IYF4 A0A1S3KGP8 A0A3B3HCC2 A0A3Q4HKX7 A0A3B3CMJ3 I3JFZ6 A0A3P9KT00 A0A3P8RDI6 H3BD84 A0A2U9BCK2 V9K9N8 G1NDI8 A0A3B4FH49 A0A3Q2VLB3 A0A3Q3NAA6 B4LNV8 A0A3Q3N858 Q291L5 H2V525 A0A3B5AVG4 A0A0P7XFA4 A0A3S2UMX4 A0A3P9AT73 A0A2D0RV59 A0A1W5A662 A0A1A8RRU8 A0A1A8QD73 A0A3Q7R1J6 A0A3Q1EX73 A0A3Q7PVY4 A0A0F8AZG5 B4MRE0 A0A1V4KED0 A0A1A8EBW2 A0A1A8KLQ0 A0A3P8XZT3 A0A1A8BD97 A0A1A8V5M8 A0A3P8TCH4
Pubmed
26354079
19121390
28756777
22118469
24845553
28004739
+ More
26823975 25474469 27129103 26334808 17510324 26483478 20566863 24508170 18362917 19820115 21347285 21282665 20966253 20920257 23761445 12364791 14747013 17210077 23537049 25244985 23254933 24487278 20798317 23258410 24495485 17554307 25186727 29451363 9215903 24402279 20838655 17994087 15632085 21551351 25835551 25069045
26823975 25474469 27129103 26334808 17510324 26483478 20566863 24508170 18362917 19820115 21347285 21282665 20966253 20920257 23761445 12364791 14747013 17210077 23537049 25244985 23254933 24487278 20798317 23258410 24495485 17554307 25186727 29451363 9215903 24402279 20838655 17994087 15632085 21551351 25835551 25069045
EMBL
NWSH01001086
PCG72702.1
RSAL01000146
RVE45912.1
KQ460313
KPJ16108.1
+ More
KQ459595 KPI95878.1 BABH01033288 KZ149909 PZC78185.1 AGBW02012034 OWR45767.1 ODYU01009571 SOQ54093.1 NEVH01022638 PNF18555.1 KK852747 KDR17236.1 GEZM01033166 JAV84383.1 GEZM01033165 JAV84384.1 GDHC01020876 GDHC01013165 JAP97752.1 JAQ05464.1 GBBI01003107 JAC15605.1 GFTR01007805 JAW08621.1 GDKW01000317 JAI56278.1 GECL01001166 JAP04958.1 GBGD01000666 JAC88223.1 CH477351 EAT42871.1 GEDC01015039 JAS22259.1 JXUM01093640 JXUM01093641 KQ564070 KXJ72841.1 DS235822 EEB17764.1 KK107054 EZA61591.1 KQ971311 EEZ99036.1 LBMM01000794 KMQ97480.1 UFQS01001365 UFQT01001365 SSX10531.1 SSX30217.1 ADTU01007009 AXCM01005936 GL766116 EFZ15575.1 KQ982580 KYQ54471.1 KQ981523 KYN40342.1 ADMH02001169 ETN63850.1 GANO01000781 JAB59090.1 AAAB01008846 EAA06302.6 GBYB01002887 JAG72654.1 APCN01004006 KQ435778 KOX74836.1 GGFJ01002878 MBW52019.1 KQ434801 KZC05878.1 KB632258 ERL90819.1 GFDL01008136 JAV26909.1 AMQN01005883 KB297068 ELU10993.1 CVRI01000057 CRL02502.1 GL445305 EFN89864.1 KB030484 ELK16055.1 GAMC01005955 GAMC01005952 GAMC01005949 GAMC01005946 JAC00610.1 GABZ01004741 JAA48784.1 OUUW01000001 SPP72807.1 AERX01007905 AFYH01004930 AFYH01004931 AFYH01004932 AFYH01004933 AFYH01004934 AFYH01004935 AFYH01004936 AFYH01004937 AFYH01004938 AFYH01004939 CP026247 AWP01519.1 JW862298 AFO94815.1 CH940648 EDW61127.1 CM000071 EAL25097.1 JARO02001621 KPP74896.1 CM012439 RVE74461.1 HAEI01000379 HAEH01018922 SBS08442.1 HAEF01003093 HAEG01012103 SBR91471.1 KQ042211 KKF17835.1 CH963850 EDW74679.1 LSYS01003385 OPJ82850.1 HAEA01014104 SBQ42584.1 HAED01003860 HAEE01013096 SBR33146.1 HADZ01000675 SBP64616.1 HAEJ01014091 SBS54548.1
KQ459595 KPI95878.1 BABH01033288 KZ149909 PZC78185.1 AGBW02012034 OWR45767.1 ODYU01009571 SOQ54093.1 NEVH01022638 PNF18555.1 KK852747 KDR17236.1 GEZM01033166 JAV84383.1 GEZM01033165 JAV84384.1 GDHC01020876 GDHC01013165 JAP97752.1 JAQ05464.1 GBBI01003107 JAC15605.1 GFTR01007805 JAW08621.1 GDKW01000317 JAI56278.1 GECL01001166 JAP04958.1 GBGD01000666 JAC88223.1 CH477351 EAT42871.1 GEDC01015039 JAS22259.1 JXUM01093640 JXUM01093641 KQ564070 KXJ72841.1 DS235822 EEB17764.1 KK107054 EZA61591.1 KQ971311 EEZ99036.1 LBMM01000794 KMQ97480.1 UFQS01001365 UFQT01001365 SSX10531.1 SSX30217.1 ADTU01007009 AXCM01005936 GL766116 EFZ15575.1 KQ982580 KYQ54471.1 KQ981523 KYN40342.1 ADMH02001169 ETN63850.1 GANO01000781 JAB59090.1 AAAB01008846 EAA06302.6 GBYB01002887 JAG72654.1 APCN01004006 KQ435778 KOX74836.1 GGFJ01002878 MBW52019.1 KQ434801 KZC05878.1 KB632258 ERL90819.1 GFDL01008136 JAV26909.1 AMQN01005883 KB297068 ELU10993.1 CVRI01000057 CRL02502.1 GL445305 EFN89864.1 KB030484 ELK16055.1 GAMC01005955 GAMC01005952 GAMC01005949 GAMC01005946 JAC00610.1 GABZ01004741 JAA48784.1 OUUW01000001 SPP72807.1 AERX01007905 AFYH01004930 AFYH01004931 AFYH01004932 AFYH01004933 AFYH01004934 AFYH01004935 AFYH01004936 AFYH01004937 AFYH01004938 AFYH01004939 CP026247 AWP01519.1 JW862298 AFO94815.1 CH940648 EDW61127.1 CM000071 EAL25097.1 JARO02001621 KPP74896.1 CM012439 RVE74461.1 HAEI01000379 HAEH01018922 SBS08442.1 HAEF01003093 HAEG01012103 SBR91471.1 KQ042211 KKF17835.1 CH963850 EDW74679.1 LSYS01003385 OPJ82850.1 HAEA01014104 SBQ42584.1 HAED01003860 HAEE01013096 SBR33146.1 HADZ01000675 SBP64616.1 HAEJ01014091 SBS54548.1
Proteomes
UP000218220
UP000283053
UP000053240
UP000053268
UP000005204
UP000007151
+ More
UP000235965 UP000027135 UP000008820 UP000069940 UP000249989 UP000009046 UP000053097 UP000007266 UP000075880 UP000036403 UP000075900 UP000075920 UP000075885 UP000075901 UP000005205 UP000075883 UP000075809 UP000078541 UP000075882 UP000069272 UP000000673 UP000075903 UP000007062 UP000076407 UP000075840 UP000053105 UP000005203 UP000076502 UP000030742 UP000076408 UP000014760 UP000183832 UP000265120 UP000008237 UP000010552 UP000265200 UP000268350 UP000085678 UP000001038 UP000261580 UP000261560 UP000005207 UP000265180 UP000265100 UP000008672 UP000246464 UP000001645 UP000261460 UP000264840 UP000261640 UP000008792 UP000001819 UP000005226 UP000261400 UP000034805 UP000265160 UP000221080 UP000192224 UP000286641 UP000257200 UP000007798 UP000190648 UP000265140 UP000265080
UP000235965 UP000027135 UP000008820 UP000069940 UP000249989 UP000009046 UP000053097 UP000007266 UP000075880 UP000036403 UP000075900 UP000075920 UP000075885 UP000075901 UP000005205 UP000075883 UP000075809 UP000078541 UP000075882 UP000069272 UP000000673 UP000075903 UP000007062 UP000076407 UP000075840 UP000053105 UP000005203 UP000076502 UP000030742 UP000076408 UP000014760 UP000183832 UP000265120 UP000008237 UP000010552 UP000265200 UP000268350 UP000085678 UP000001038 UP000261580 UP000261560 UP000005207 UP000265180 UP000265100 UP000008672 UP000246464 UP000001645 UP000261460 UP000264840 UP000261640 UP000008792 UP000001819 UP000005226 UP000261400 UP000034805 UP000265160 UP000221080 UP000192224 UP000286641 UP000257200 UP000007798 UP000190648 UP000265140 UP000265080
PRIDE
Interpro
Gene 3D
ProteinModelPortal
A0A2A4JMS1
A0A3S2LG08
A0A194RE51
A0A194PSD4
H9JN62
A0A2W1BX13
+ More
A0A212EWC9 A0A2H1WLY5 A0A2J7PQE6 A0A067REC1 A0A1Y1MFF0 A0A1Y1MF67 A0A146KMH8 A0A023F2Q0 A0A224XKR0 A0A0P4W4K8 A0A0V0GAA6 A0A069DVV8 Q0IFF4 A0A1B6D987 A0A1S4FBE3 A0A182GWM5 E0VWK8 A0A026X0R1 D6WCJ9 A0A182J5K6 A0A0J7NYK0 A0A336L2W6 A0A182R6F5 A0A182W3Y6 A0A182PFE8 A0A182T8P1 A0A158P279 A0A182MRP2 E9IV14 A0A151X210 A0A195FJA5 A0A182KJZ3 A0A182FJC8 W5JLS5 A0A182UYB6 U5EZH6 Q7PS14 A0A182XPP4 A0A0C9QQG9 A0A182HIM8 A0A0M9A227 A0A2M4BG34 A0A087ZZZ9 A0A154P3R5 U4UA52 A0A182YCN9 A0A1Q3FHA4 R7UWJ4 A0A1J1ITR8 A0A3P8UQE7 E2B382 L5KY03 W8CAA2 A0A3P9HQH6 K9J2H1 A0A3B0IYF4 A0A1S3KGP8 A0A3B3HCC2 A0A3Q4HKX7 A0A3B3CMJ3 I3JFZ6 A0A3P9KT00 A0A3P8RDI6 H3BD84 A0A2U9BCK2 V9K9N8 G1NDI8 A0A3B4FH49 A0A3Q2VLB3 A0A3Q3NAA6 B4LNV8 A0A3Q3N858 Q291L5 H2V525 A0A3B5AVG4 A0A0P7XFA4 A0A3S2UMX4 A0A3P9AT73 A0A2D0RV59 A0A1W5A662 A0A1A8RRU8 A0A1A8QD73 A0A3Q7R1J6 A0A3Q1EX73 A0A3Q7PVY4 A0A0F8AZG5 B4MRE0 A0A1V4KED0 A0A1A8EBW2 A0A1A8KLQ0 A0A3P8XZT3 A0A1A8BD97 A0A1A8V5M8 A0A3P8TCH4
A0A212EWC9 A0A2H1WLY5 A0A2J7PQE6 A0A067REC1 A0A1Y1MFF0 A0A1Y1MF67 A0A146KMH8 A0A023F2Q0 A0A224XKR0 A0A0P4W4K8 A0A0V0GAA6 A0A069DVV8 Q0IFF4 A0A1B6D987 A0A1S4FBE3 A0A182GWM5 E0VWK8 A0A026X0R1 D6WCJ9 A0A182J5K6 A0A0J7NYK0 A0A336L2W6 A0A182R6F5 A0A182W3Y6 A0A182PFE8 A0A182T8P1 A0A158P279 A0A182MRP2 E9IV14 A0A151X210 A0A195FJA5 A0A182KJZ3 A0A182FJC8 W5JLS5 A0A182UYB6 U5EZH6 Q7PS14 A0A182XPP4 A0A0C9QQG9 A0A182HIM8 A0A0M9A227 A0A2M4BG34 A0A087ZZZ9 A0A154P3R5 U4UA52 A0A182YCN9 A0A1Q3FHA4 R7UWJ4 A0A1J1ITR8 A0A3P8UQE7 E2B382 L5KY03 W8CAA2 A0A3P9HQH6 K9J2H1 A0A3B0IYF4 A0A1S3KGP8 A0A3B3HCC2 A0A3Q4HKX7 A0A3B3CMJ3 I3JFZ6 A0A3P9KT00 A0A3P8RDI6 H3BD84 A0A2U9BCK2 V9K9N8 G1NDI8 A0A3B4FH49 A0A3Q2VLB3 A0A3Q3NAA6 B4LNV8 A0A3Q3N858 Q291L5 H2V525 A0A3B5AVG4 A0A0P7XFA4 A0A3S2UMX4 A0A3P9AT73 A0A2D0RV59 A0A1W5A662 A0A1A8RRU8 A0A1A8QD73 A0A3Q7R1J6 A0A3Q1EX73 A0A3Q7PVY4 A0A0F8AZG5 B4MRE0 A0A1V4KED0 A0A1A8EBW2 A0A1A8KLQ0 A0A3P8XZT3 A0A1A8BD97 A0A1A8V5M8 A0A3P8TCH4
PDB
1ON8
E-value=5.27638e-28,
Score=312
Ontologies
PATHWAY
GO
GO:0015012
GO:0016021
GO:0015020
GO:0006486
GO:0006024
GO:0050508
GO:0009058
GO:0016757
GO:0030170
GO:0050650
GO:0001649
GO:0045743
GO:0048538
GO:0035118
GO:0051216
GO:0031290
GO:0048920
GO:0030516
GO:0008543
GO:0042476
GO:0097374
GO:0030198
GO:0001707
GO:0005794
GO:0046982
GO:0042803
GO:0043541
GO:0030154
GO:0008375
GO:0001503
GO:0015014
GO:0050509
GO:0045880
GO:0090098
GO:0030206
GO:0005783
GO:0030210
GO:0008101
GO:0090263
GO:0005887
GO:0016907
GO:0019538
GO:0030145
GO:0003676
GO:0016491
GO:0015930
PANTHER
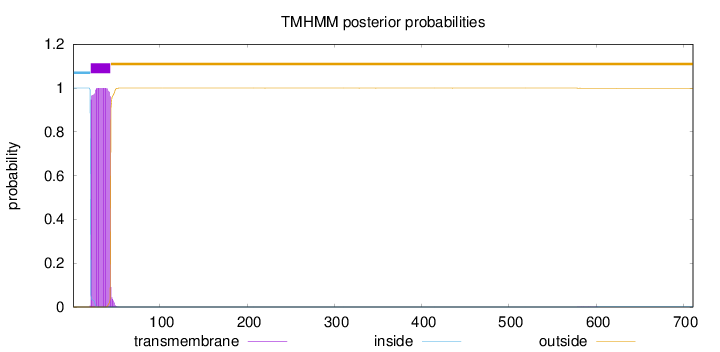
Topology
Length:
711
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.9972899999999
Exp number, first 60 AAs:
22.97931
Total prob of N-in:
0.99984
POSSIBLE N-term signal
sequence
inside
1 - 20
TMhelix
21 - 43
outside
44 - 711
Population Genetic Test Statistics
Pi
207.426686
Theta
174.144339
Tajima's D
0.374067
CLR
1.089992
CSRT
0.47647617619119
Interpretation
Uncertain
