Gene
KWMTBOMO13577
Pre Gene Modal
BGIBMGA001382
Annotation
PREDICTED:_serum_response_factor_homolog_[Papilio_xuthus]
Transcription factor
Location in the cell
Nuclear Reliability : 4.54
Sequence
CDS
ATGGACGCGCCCGGCGGAGCGCGGGAGTCCCGCTTCGGGCCGTCTTACATGGGGTTGTTGGGGGAGATGCCGGACATGTACAGCGCGGGCAGGCCTCCGAGCGCGCTCGGCGGTGCTGGCCCCTTGCGGCCCGGCATGCAACCTTGCCCGATGCCCAGGGCTGGAGTCAAACGTCCATCGGACCAGTGCTACGATGATAGACCATCACAAAGCGTTCCCCTCGACCACTGCCCAATGCCGGACATAGCTGATGATGGTTATGCCTCCCTGCAACCCAAGAAGTCACCCCCTTCTAATGGCAAAAAGACTAAAGGTCGCGTCAAGATCAAAATGGAGTATATAGATAATAAACTGCGACGGTACACAACTTTCTCCAAGCGGAAGACTGGGATTATGAAGAAGGCATACGAGCTATCCACTTTGACGGGGACACAAGTGATGTTGCTGGTCGCGTCGGAGACCGGCCACGTGTATACGTTCGCGACGCGGAAACTTCAGCCCATGATCACGTCCGATTCTGGGAAAAGGCTGATCCAGACGTGCCTTAACTCACCAGACCCGCCCACGACCAGCGAACAGCGCATGGCGGCTACTGGCTTCGAGGAGACCGAGCTCACGTATAACGTTGTAGACGAGGATATGAAGGTGAGGCAATTGGCATACGCGGGCACCGCGCAGTACCCTATAGAGCACCATCCAGGGCTGGCCCCGTCGCCGCTGCAGCAGTACCATCAGCACCCACCCTGTCCGTCACCGCTGCCCCTCAGCTCGCTGGGGCAGCCCTATTCTCACGCTCATCTATCTCACCCCCACATGTCGCATCATCCTCAGCGGTAG
Protein
MDAPGGARESRFGPSYMGLLGEMPDMYSAGRPPSALGGAGPLRPGMQPCPMPRAGVKRPSDQCYDDRPSQSVPLDHCPMPDIADDGYASLQPKKSPPSNGKKTKGRVKIKMEYIDNKLRRYTTFSKRKTGIMKKAYELSTLTGTQVMLLVASETGHVYTFATRKLQPMITSDSGKRLIQTCLNSPDPPTTSEQRMAATGFEETELTYNVVDEDMKVRQLAYAGTAQYPIEHHPGLAPSPLQQYHQHPPCPSPLPLSSLGQPYSHAHLSHPHMSHHPQR
Summary
Uniprot
A0A2A4JMP3
A0A212EI12
A0A2W1BSY0
A0A0H3UYK2
A0A1Y1LB27
C0KZ12
+ More
D6WCK0 A0A1L8DI72 V5IA35 E0VYQ3 A0A1Y1LC57 A0A067R4V2 A0A1S3DVS1 A0A0A9ZAQ6 A0A2A3EB75 A0A154PQ61 A0A310SMG9 A0A2J7PQE2 A0A0L7R2V5 A0A146M887 A0A1I8M8Y2 A0A146LKN9 A0A0K8T2J3 A0A1W4WAC6 A0A146KVU2 A0A0A9YH03 A0A0N0U405 A0A0K8T2Y2 A0A0L0CF08 A0A1B6LRR8 A0A1B6G2L2 A0A088A7B8 A0A0P5IMP3 A0A0P5HK91 A0A0D6DQH9 A0A1I8M8X5 A0A0P5UPT6 A0A0P5E600 T1PH56 A0A0P4XLQ3 A0A0C9R2T8 A0A158NTW2 A0A195FPH9 A0A1B6LY53 A0A1B6K564 A0A195CTS3 A0A0P4ZDE2 A0A0C9RPV3 A0A0P5SNK0 A0A0P6DHQ9 A0A0P4Z8X3 A0A2S2P4J6 A0A0P5D142
D6WCK0 A0A1L8DI72 V5IA35 E0VYQ3 A0A1Y1LC57 A0A067R4V2 A0A1S3DVS1 A0A0A9ZAQ6 A0A2A3EB75 A0A154PQ61 A0A310SMG9 A0A2J7PQE2 A0A0L7R2V5 A0A146M887 A0A1I8M8Y2 A0A146LKN9 A0A0K8T2J3 A0A1W4WAC6 A0A146KVU2 A0A0A9YH03 A0A0N0U405 A0A0K8T2Y2 A0A0L0CF08 A0A1B6LRR8 A0A1B6G2L2 A0A088A7B8 A0A0P5IMP3 A0A0P5HK91 A0A0D6DQH9 A0A1I8M8X5 A0A0P5UPT6 A0A0P5E600 T1PH56 A0A0P4XLQ3 A0A0C9R2T8 A0A158NTW2 A0A195FPH9 A0A1B6LY53 A0A1B6K564 A0A195CTS3 A0A0P4ZDE2 A0A0C9RPV3 A0A0P5SNK0 A0A0P6DHQ9 A0A0P4Z8X3 A0A2S2P4J6 A0A0P5D142
Pubmed
EMBL
NWSH01001086
PCG72703.1
AGBW02014734
OWR41122.1
KZ149909
PZC78182.1
+ More
KJ158242 AIZ67912.1 GEZM01063287 JAV69135.1 FJ647802 ACN43332.1 KQ971311 EEZ99037.2 GFDF01007951 JAV06133.1 GALX01001875 JAB66591.1 DS235846 EEB18509.1 GEZM01063286 JAV69136.1 KK852747 KDR17235.1 GBHO01002105 GBRD01006068 JAG41499.1 JAG59753.1 KZ288296 PBC28940.1 KQ435012 KZC13857.1 KQ762343 OAD55936.1 NEVH01022638 PNF18552.1 KQ414664 KOC65210.1 GDHC01003849 JAQ14780.1 GDHC01010610 JAQ08019.1 GBRD01006069 JAG59752.1 GDHC01018018 JAQ00611.1 GBHO01012698 GBRD01006067 JAG30906.1 JAG59754.1 KQ435866 KOX70294.1 GBRD01006070 GDHC01005615 JAG59751.1 JAQ13014.1 JRES01000476 KNC30983.1 GEBQ01013611 JAT26366.1 GECZ01013094 JAS56675.1 GDIQ01210551 JAK41174.1 GDIQ01228284 JAK23441.1 HF912428 CCV01216.1 GDIQ01103184 GDIP01127488 JAL48542.1 JAL76226.1 GDIP01151099 JAJ72303.1 KA648106 AFP62735.1 GDIP01240792 JAI82609.1 GBYB01010519 JAG80286.1 ADTU01026164 ADTU01026165 KQ981382 KYN42306.1 GEBQ01011348 JAT28629.1 GECU01001403 JAT06304.1 KQ977279 KYN04093.1 GDIP01214234 JAJ09168.1 GBYB01010520 JAG80287.1 GDIP01137489 LRGB01000781 JAL66225.1 KZS15922.1 GDIQ01080401 JAN14336.1 GDIP01216082 JAJ07320.1 GGMR01011519 MBY24138.1 GDIP01164336 JAJ59066.1
KJ158242 AIZ67912.1 GEZM01063287 JAV69135.1 FJ647802 ACN43332.1 KQ971311 EEZ99037.2 GFDF01007951 JAV06133.1 GALX01001875 JAB66591.1 DS235846 EEB18509.1 GEZM01063286 JAV69136.1 KK852747 KDR17235.1 GBHO01002105 GBRD01006068 JAG41499.1 JAG59753.1 KZ288296 PBC28940.1 KQ435012 KZC13857.1 KQ762343 OAD55936.1 NEVH01022638 PNF18552.1 KQ414664 KOC65210.1 GDHC01003849 JAQ14780.1 GDHC01010610 JAQ08019.1 GBRD01006069 JAG59752.1 GDHC01018018 JAQ00611.1 GBHO01012698 GBRD01006067 JAG30906.1 JAG59754.1 KQ435866 KOX70294.1 GBRD01006070 GDHC01005615 JAG59751.1 JAQ13014.1 JRES01000476 KNC30983.1 GEBQ01013611 JAT26366.1 GECZ01013094 JAS56675.1 GDIQ01210551 JAK41174.1 GDIQ01228284 JAK23441.1 HF912428 CCV01216.1 GDIQ01103184 GDIP01127488 JAL48542.1 JAL76226.1 GDIP01151099 JAJ72303.1 KA648106 AFP62735.1 GDIP01240792 JAI82609.1 GBYB01010519 JAG80286.1 ADTU01026164 ADTU01026165 KQ981382 KYN42306.1 GEBQ01011348 JAT28629.1 GECU01001403 JAT06304.1 KQ977279 KYN04093.1 GDIP01214234 JAJ09168.1 GBYB01010520 JAG80287.1 GDIP01137489 LRGB01000781 JAL66225.1 KZS15922.1 GDIQ01080401 JAN14336.1 GDIP01216082 JAJ07320.1 GGMR01011519 MBY24138.1 GDIP01164336 JAJ59066.1
Proteomes
Pfam
PF00319 SRF-TF
SUPFAM
SSF55455
SSF55455
Gene 3D
ProteinModelPortal
A0A2A4JMP3
A0A212EI12
A0A2W1BSY0
A0A0H3UYK2
A0A1Y1LB27
C0KZ12
+ More
D6WCK0 A0A1L8DI72 V5IA35 E0VYQ3 A0A1Y1LC57 A0A067R4V2 A0A1S3DVS1 A0A0A9ZAQ6 A0A2A3EB75 A0A154PQ61 A0A310SMG9 A0A2J7PQE2 A0A0L7R2V5 A0A146M887 A0A1I8M8Y2 A0A146LKN9 A0A0K8T2J3 A0A1W4WAC6 A0A146KVU2 A0A0A9YH03 A0A0N0U405 A0A0K8T2Y2 A0A0L0CF08 A0A1B6LRR8 A0A1B6G2L2 A0A088A7B8 A0A0P5IMP3 A0A0P5HK91 A0A0D6DQH9 A0A1I8M8X5 A0A0P5UPT6 A0A0P5E600 T1PH56 A0A0P4XLQ3 A0A0C9R2T8 A0A158NTW2 A0A195FPH9 A0A1B6LY53 A0A1B6K564 A0A195CTS3 A0A0P4ZDE2 A0A0C9RPV3 A0A0P5SNK0 A0A0P6DHQ9 A0A0P4Z8X3 A0A2S2P4J6 A0A0P5D142
D6WCK0 A0A1L8DI72 V5IA35 E0VYQ3 A0A1Y1LC57 A0A067R4V2 A0A1S3DVS1 A0A0A9ZAQ6 A0A2A3EB75 A0A154PQ61 A0A310SMG9 A0A2J7PQE2 A0A0L7R2V5 A0A146M887 A0A1I8M8Y2 A0A146LKN9 A0A0K8T2J3 A0A1W4WAC6 A0A146KVU2 A0A0A9YH03 A0A0N0U405 A0A0K8T2Y2 A0A0L0CF08 A0A1B6LRR8 A0A1B6G2L2 A0A088A7B8 A0A0P5IMP3 A0A0P5HK91 A0A0D6DQH9 A0A1I8M8X5 A0A0P5UPT6 A0A0P5E600 T1PH56 A0A0P4XLQ3 A0A0C9R2T8 A0A158NTW2 A0A195FPH9 A0A1B6LY53 A0A1B6K564 A0A195CTS3 A0A0P4ZDE2 A0A0C9RPV3 A0A0P5SNK0 A0A0P6DHQ9 A0A0P4Z8X3 A0A2S2P4J6 A0A0P5D142
PDB
1SRS
E-value=2.01813e-39,
Score=406
Ontologies
GO
Topology
Subcellular location
Nucleus
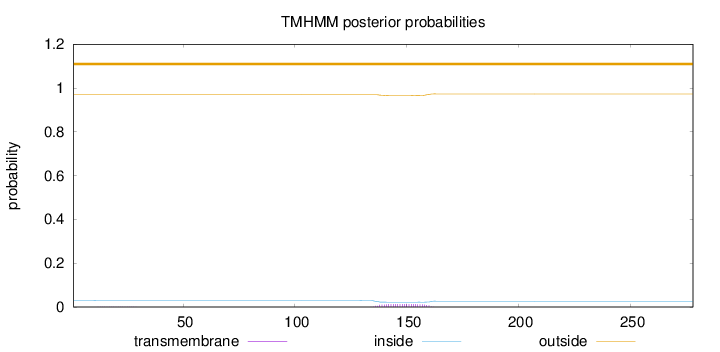
Length:
278
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.29635
Exp number, first 60 AAs:
0.00328
Total prob of N-in:
0.03049
outside
1 - 278
Population Genetic Test Statistics
Pi
318.439255
Theta
203.128507
Tajima's D
1.348008
CLR
0.196753
CSRT
0.749562521873906
Interpretation
Uncertain
